INTRODUCTION
In Canada, human salmonellosis is notifiable, with about 5000 cases reported annually, second only to campylobacteriosis in causing bacterial gastroenteritis [1]. The epidemiology of human non-typhoidal salmonellosis is complex involving various animal reservoirs and several transmission pathways, the most important of which is foodborne. Several serovars also cause disease in animal species. Of note, two reportable host-specific serovars (S. Gallinarum and S. Pullorum) cause severe disease in poultry [2]. Additionally, the detection of antimicrobial resistance in Salmonella increases both human and animal health concerns. Because of the burden of human salmonellosis, its zoonotic and foodborne epidemiology, and the impact of Salmonella infections on the health of animals and the sustainability of animal livestock industries, Canada has long-standing surveillance systems, with specific objectives to monitor Salmonella in humans, food animals, and food.
Salmonella serovar Enteritidis emerged as a prevalent cause of human salmonellosis in Canada and globally in the 1980s [Reference Rodriguez, Tauxe and Rowe3]. S. Enteritidis has been ranked in the top three non-typhoidal serovars, ranging from 12% to 27% of all Salmonella infections reported in Canada from 1999 to 2006 [4]. Large outbreaks since 2005 (see Results section), and increasing rates of illness overall [4], have highlighted S. Enteritidis as an emerging pathogen in Canada causing a significant burden of illness. This paper provides a comprehensive picture of S. Enteritidis in Canada from 2003 to 2009 utilizing available data on Salmonella from national and targeted animal and human surveillance programmes. The specific objectives were to describe the trends of S. Enteritidis in humans and the agri-food sector and to explore the potential driving causes of national trends.
METHODS
Data sources
National surveillance data
The National Enteric Surveillance Program (NESP) provides weekly aggregate reports of laboratory-confirmed enteric disease cases reported by the provincial public health authorities. The purpose of NESP is to detect outbreaks and report national trends (see http://www.nml-lnm.gc.ca/NESP-PNSME/index-eng.htm for details).
Laboratory-confirmed human salmonellosis isolates are submitted to the National Microbiology Laboratory (NML) as part of reference requests, active and passive surveillance programmes, surveys or outbreak and cluster investigations. Provincial laboratories perform pulsed-field gel electrophoresis (PFGE) using standardized PulseNet Canada protocols [Reference Ribot5] and submit the resulting data to the NML for PFGE pattern designation. Submitted isolates are phage-typed at the NML using the international S. Enteritidis phage-typing scheme [Reference Ward, De Sa and Rowe6], and for select provincial microbiology laboratories, PFGE and serotyping are also performed.
Targeted surveillance data
The Canadian Integrated Program for Antimicrobial Resistance Surveillance (CIPARS) monitors antimicrobial use and antimicrobial resistance in selected species of enteric bacteria, including Salmonella, from humans, animals and animal-derived food sources across Canada (see http://www.phac-aspc.gc.ca/cipars-picra/surv-eng.php for details). Human Salmonella isolates are submitted by provincial public health laboratories to NML for further subtyping and antimicrobial susceptibility testing. Data on Salmonella from the agri-food sectors are collected for various commodities: pigs on farm, caecal samples from slaughtered chickens and pigs, and retail chicken and pork meats. All Salmonella isolates recovered from animals and food are forwarded to the Salmonella Typing Laboratory (STL) of the Laboratory for Food-borne Zoonoses, Public Health Agency of Canada (PHAC), to have serotype, phage type, and antimicrobial susceptibility determined. Some Salmonella isolates from diagnostic animal samples submitted by veterinarians and from various government monitoring programmes, such as the testing of fluff from approved hatchery supplier flocks [7] undergo the same testing as the CIPARS isolates described above. However, isolates from government monitoring programmes do not routinely undergo antimicrobial susceptibility testing.
C-EnterNet is an integrated sentinel site surveillance programme that systematically samples and tests human enteric pathogens from three exposure sources [retail food (chicken, pork, beef), on-farm (broiler chicken, swine, dairy, beef), and untreated surface water] in parallel with human enteric pathogen laboratory-based surveillance and enhanced case follow-up on exposures (see http://www.phac-aspc.gc.ca/c-enternet/index-eng.php for details). The programme was implemented in June 2005 at the pilot sentinel site (Waterloo Region, Ontario): a population of about 500 000 residents. The enhanced follow-up of all human cases enables classification of cases as international travel-related, outbreak-related or endemic sporadic cases.
Outbreak data
Outbreak data are not captured in a standardized or systematic manner across the country. S. Enteritidis outbreaks in Canada during the study period were retrieved through (1) a peer-reviewed literature search limited to Canadian outbreaks from 2003–2009 conducted in PubMed using the key words: Salmonella, Enteritidis, and outbreak; (2) a review of investigation records documented by the Outbreak Management Division at the Centre for Food-borne, Environmental and Zoonotic Infectious Diseases, PHAC; and (3) provincial outbreak investigation records requested from provincial and territorial partners.
Data analysis
Data collected from national and targeted animal and human surveillance programmes from January 2003 to December 2009 were analysed. Annual incidence rates for human salmonellosis were computed using the national and sentinel site population estimates [8]. Temporal trends (annual and seasonal) were analysed based on monthly counts using a negative binominal regression model for all cases and for the six most frequent phage types separately. Cases were described by age, sex, phage type, PFGE patterns and antimicrobial susceptibility. S. Enteritidis isolated from non-human samples were described by phage type, antimicrobial susceptibility, PFGE pattern and trends by food commodity and data source. To test statistical differences, when appropriate, logistic regression analysis was conducted using SAS version 9.1 (SAS Institute Inc., USA) and the χ2 test and critical ratio (z) test using EpiCalc2000 version 1.02 [Reference Gilman and Myatt9]. All statistical significant differences were identified by using a P value threshold of 0·05.
RESULTS
S. Enteritidis in humans
Overall incidence
A total of 10 616 laboratory-confirmed S. Enteritidis human cases were reported in Canada from 2003 to 2009 via NESP. The national annual incidence rate increased from 2·16/100 000 person-years in 2003 to 5·79/100 000 in 2009 (63% increase), while the incidence rate for all non-typhoidal Salmonella infections increased from 16·29/100 000 person-years in 2003 to 18·03/100 000 in 2009 (Fig. 1) (10% increase). Of all reported Salmonella, the proportion of S. Enteritidis isolates rose from 12·7% in 2003 to 32·1% in 2009 (Fig. 1). The increase of S. Enteritidis cases was observed in each of the 10 provinces and three territories, with varying incidence rates (Fig. 2) and distribution of genetic subtypes (see Microbial features section below).
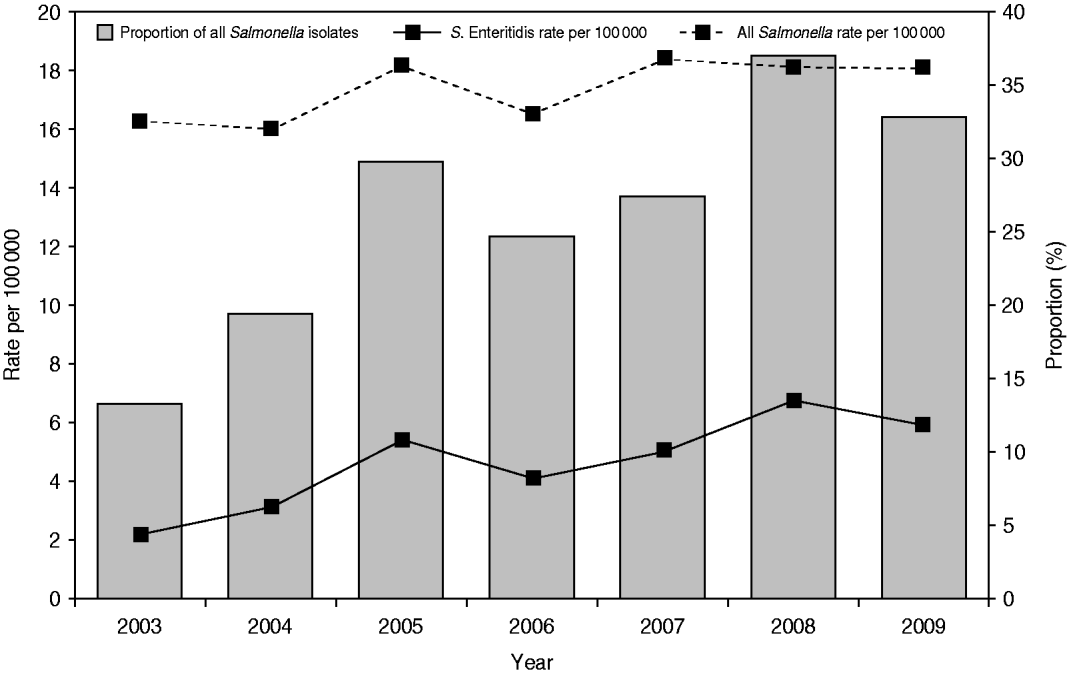
Fig. 1. Laboratory-confirmed Salmonella Enteritidis case incidence rate (per 100 000) and proportion among all non-typhoidal Salmonella isolates, by year, in Canada, 2003–2009 (source: National Enteric Surveillance Program).

Fig. 2. Laboratory-confirmed Salmonella Enteritidis case incidence rate (per 100 000), by year and province/territory, in Canada, 2003–2009 (source: National Enteric Surveillance Program). YT, Yukon Territory; NT, Northwest Territory; NU, Nunavut; BC, British Columbia; AB, Alberta; SK, Saskatchewan; MB, Manitoba; ON, Ontario; QU, Québec; NB, New Brunswick; NS, Nova Scotia; PEI, Prince Edward Island; NL, Newfoundland.
Endemic sporadic cases
Nationally, it was difficult to distinguish between outbreak-related cases, international travel-related cases, and endemic sporadic, domestically acquired cases because of limited epidemiological data accompanying the isolates. At the sentinel site level (C-EnterNet) the incidence rate for endemic sporadic S. Enteritidis cases increased from 0·41/100 000 person-years in 2005 to 3·66/100 000 in 2009.
Outbreak-related cases
Six S. Enteritidis outbreaks with well documented investigations were identified from 2003 to 2009. S. Enteritidis outbreaks were largely attributed to PT8 and PT13 (Fig. 3). In 2004, a cluster of 10 cases of S. Enteritidis PT8 linked to a single restaurant in Alberta was identified. Although a food source was not identified, the resulting investigation implicated a food handler as the probable source (V. Mah, Alberta Health and Wellness, personal communication, February 2009). In 2005, a large S. Enteritidis PT13 outbreak occurred in Ontario, resulting in 552 laboratory-confirmed cases of illness, and the investigation identified mung bean sprouts as the source of infection. This outbreak was described in the peer-reviewed literature [Reference Olson10]. In 2006, an increase in S. Enteritidis PT13 occurred in Ontario resulting in an investigation that identified either chicken or eggs as a potential source of infection, although this could not be confirmed (Ontario Ministry of Health and Long-Term Care, personal communication, February 2009). In 2007, a notable increase in S. Enteritidis PT13 occurred across three provinces (Ontario, British Columbia, New Brunswick). The subsequent investigations, undertaken provincially and nationally, indicated several potential sources of infection including chicken and eggs; however, no definitive source was confirmed (Outbreak Management Division, CFEZID, personal communication, February 2009). In August 2008, an outbreak in the province of Québec involving more than 150 cases of S. Enteritidis PT13 was attributed to a pasteurized cheese made in this province (C. Gaulin, Québec Ministère de la Santé et des Services sociaux du Québec, personal communication, February 2009). British Columbia identified an increase in S. Enteritidis PT8 in June 2008. The resulting investigation identified eggs as the most likely source of infection. The increase and accompanying investigation have continued into 2011 [Reference Thomas and Wilson11].
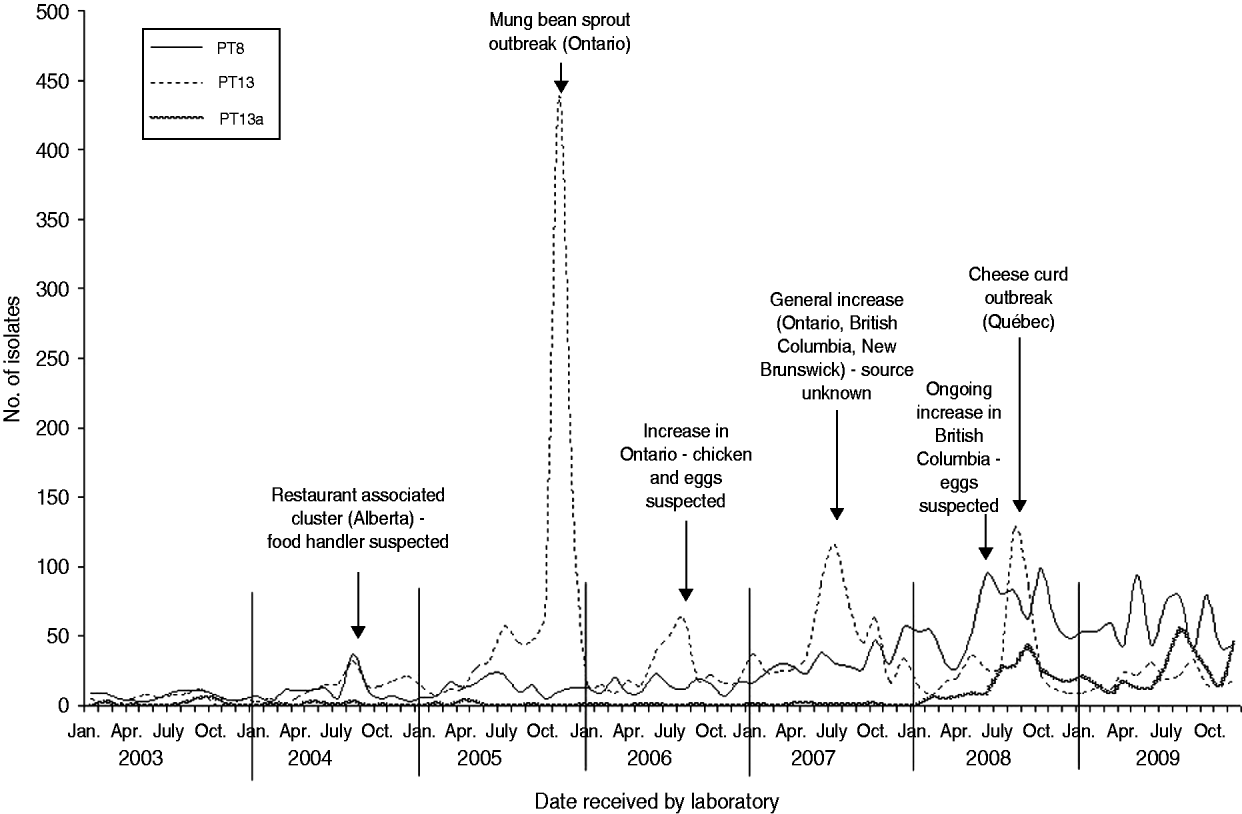
Fig. 3. Salmonella Enteritidis PT8, PT13a and PT13 isolates by month, Canada 2003–2009 (source: National Microbiology Laboratory and outbreak identifications, Outbreak Management Division, Centre for Food-borne, Environmental, and Zoonotic Infectious Diseases, Public Health Agency of Canada and Provincial Public Health Authorities).
International travel-related cases
At C-EnterNet's sentinel site, 36% (69/190) of S. Enteritidis cases reported between mid-2005 and 2009 were linked to travel outside Canada. Overall, the most frequent travel destination reported by these travel-related cases was to the Caribbean.
Age and gender distribution of cases
There was no significant difference in the gender distribution across all surveillance systems (Table 1). Nationally, CIPARS data showed that the S. Enteritidis cases were older than all Salmonella cases, with fewer cases in children aged <10 years and more cases in adults aged between 30 and 59 years (Table 1). The age distribution of the S. Enteritidis cases at the sentinel site level was similar to the national estimate, except it was higher for adults aged 20–24 years (15% and 8%, respectively) (Table 1).
Table 1. Age and gender distribution (%) of non-typhoidal Salmonella and S. Enteritidis cases, in Canada, 2003–2009 (sources: C-EnterNet and Canadian Integrated Program for Antimicrobial Resistance Surveillance (CIPARS)
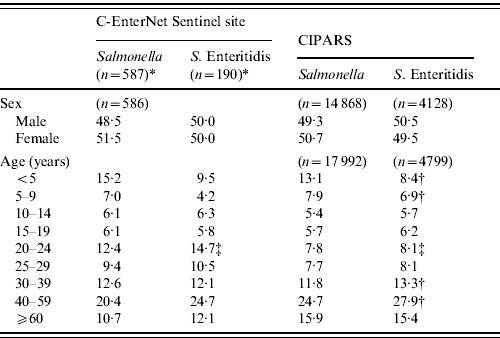
* Includes endemic, travel- and outbreak-related cases.
† Significantly different proportion of CIPARS S. Enteritidis cases than all CIPARS Salmonella serovars (P<0·05).
‡ Significantly different proportion of S.Enteritidis cases than other surveillance programme (P<0·05).
Microbial features
From 2003 to 2009, 10 555 human S. Enteritidis isolates were phage-typed by the NML. The most common phage types were PT13 (25%), PT8 (22%), PT4 (13%), and PT1 (8%) (Table 2). Prior to 2004, PT13, PT8 and PT13a represented less than 25% of all phage types. Combinations of these three phage types are presently found in most provinces, representing 62% nationally in 2009. The phage-type distribution was comparable to CIPARS data, which is a subset of 6923 isolates of the national database (Table 2). The temporal trends for the six most common phage types showed various annual and seasonal patterns (Figs 4 and 5). Overall, compared to December, significantly more S. Enteritidis cases were reported in August and September and fewer in November. Phage types 13, 13a and 8 had summer seasonal peaks, while PT1, PT4 and PT6a showed winter seasonal peaks. PT13 was associated with four large outbreaks between 2003 and 2009 (Fig. 4). With removal of the Ontario outbreak related to mung bean sprouts in 2005, PT13 significantly increased in 2004, 2005 and 2007 compared to the preceding year and decreased in 2008 and 2009. PT13 showed an increase in Ontario in 2005, 2006 and 2007; a marked increase was observed in British Columbia in 2007; and Québec had a large outbreak in 2008 associated with cheese curds. PT13 showed a seasonal peak in summer (July–September) and a drop in winter (January to March). PT8 showed an upward trend from 2003 to 2009 with a statistically significant increase from 2006 to 2007 and from 2007 to 2008. No statistically significant monthly differences were found. Between 2005 and 2006, regional differences were observed, with PT8 as the predominant phage type in the Eastern and Western provinces. Notable increases in PT8 were observed in almost all provinces since 2007; British Columbia showed a dramatic increase in 2008. PT13a increased significantly from one year to another starting in 2008 in almost all provinces and without any obvious seasonality (Fig. 4). PT4 peaked in 2006, followed by a significant decrease from 2006 to 2007 and from 2008 to 2009 with two seasonal peaks; one from January to April and another in late summer (August and September; Fig. 5). PT1 showed no obvious annual trend, but peaked seasonally from January to April (Fig. 5). Finally, PT6a significantly increased from 2003 to 2004 and from 2007 to 2008 before significantly decreasing in 2009. There was a significant seasonal drop in cases between June and August (Fig. 5).

Fig. 4. Monthly actual and predicted numbers of S. Enteritidis cases for phage types 13, 8 and 13a, in Canada, 2003–2009 (source: National Microbiology Laboratory). The predicted values are the result of modelling the data using a negative binomial regression and assist in predicting seasonality.
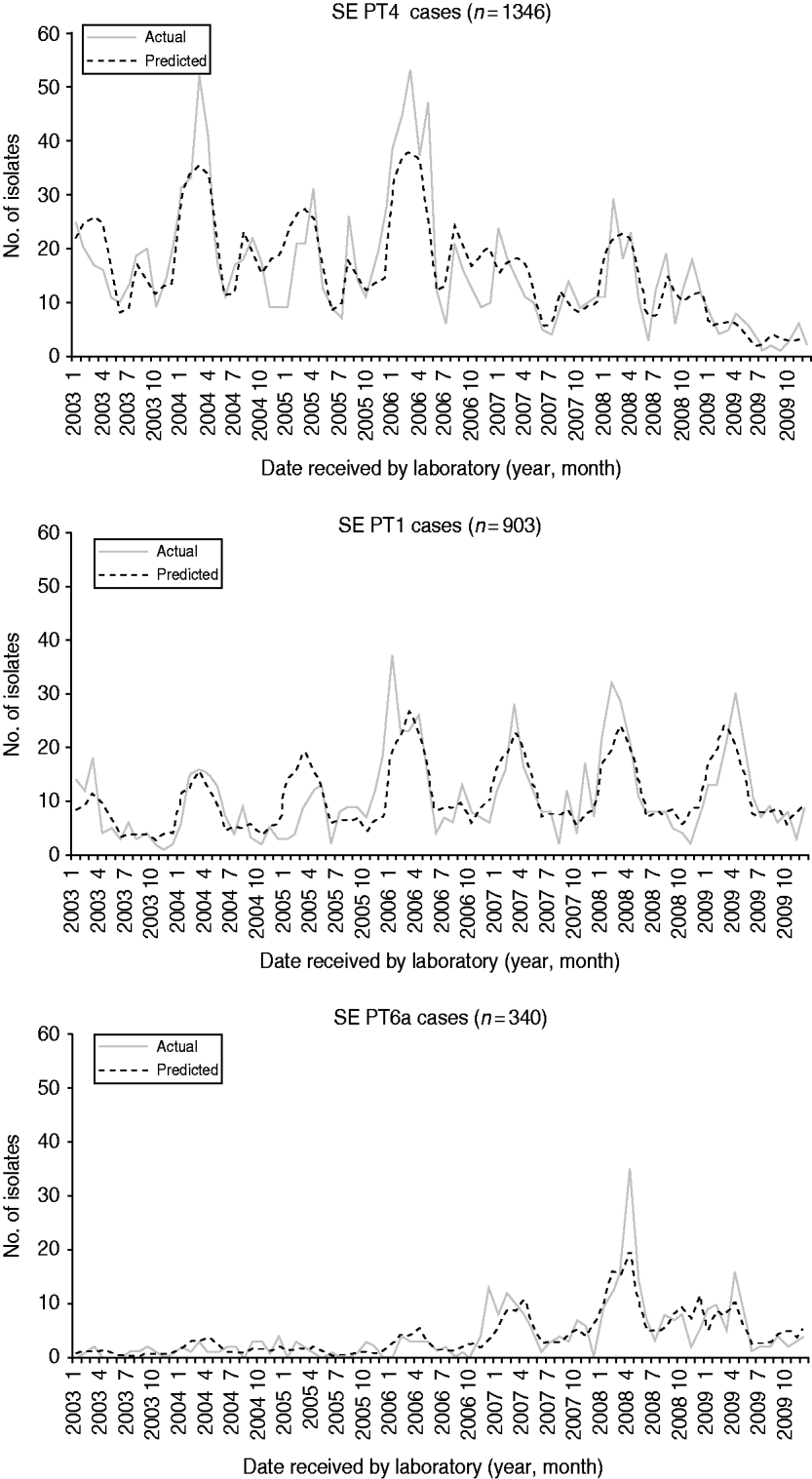
Fig. 5. Monthly actual and predicted numbers of S. Enteritidis cases for phage types 1, 4 and 6a, in Canada, 2003–2009 (source: National Microbiology Laboratory). The predicted values are the result of modelling the data using a negative binomial regression and assist in predicting seasonality.
Table 2. Top five S. Enteritidis phage types in human and non-human isolates by surveillance data sources, in Canada, 2003–2009 (sources: National Microbiology Laboratory (NML), Canadian Integrated Program for Antimicrobial Resistance Surveillance (CIPARS) and C-EnterNet)
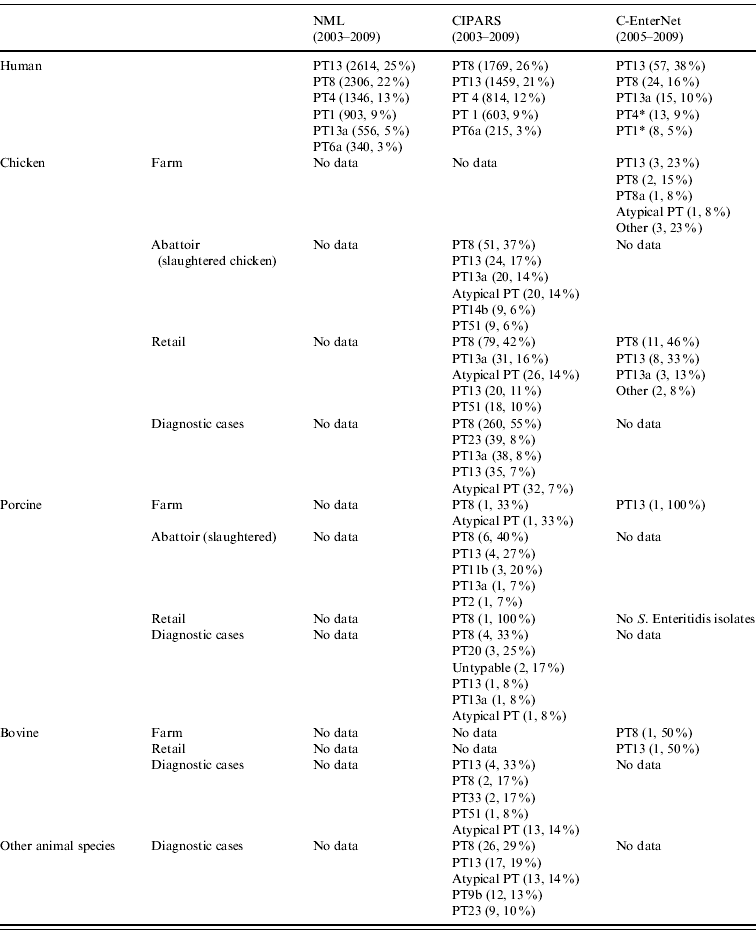
* Data from C-EnterNet indicate that these phage types are primarily associated with cases linked to international travel.
† Other animal species include: unspecified bird species (55), ducks (12), dogs (6), cats (4), rodents (3), aquatic mammals (2), geese (2), reptiles (2), turkeys (2), amphibians (1), horses (1), and mink (1).
In C-EnterNet's sentinel site, PT13 was the most common phage type (Table 2). All PT13 isolates were endemic, with the majority (40/57) associated with an outbreak in 2005. The majority of PT8 isolates (21/24) were also endemic. All PT1 isolates were travel-related and 77% of PT4 isolates were also travel-related.
In general, the predominant XbaI PFGE patterns identified by C-EnterNet with their corresponding phage types were as follows: SENXAI.0001 with PT4, SENXAI.0003 with PT8, SENXAI.0006 with PT13a and SENXAI.0038 with PT13 (Table 3). Considering isolates from both human cases and the chicken commodity, 10 (83%) of 12 PT4 isolates were SENXAI.0001, 92 (89%) of 103 PT8 isolates were SENXAI.0003, 35 (69%) of 51 PT13a isolates were SENXAI.0006 and 48 (96%) of 50 PT13 isolates were SENXAI.0038.
Table 3. Phage type and PFGE pattern for S. Enteritidis isolated from human cases, chickens, and retail chicken meat in Canada, 2005–2009
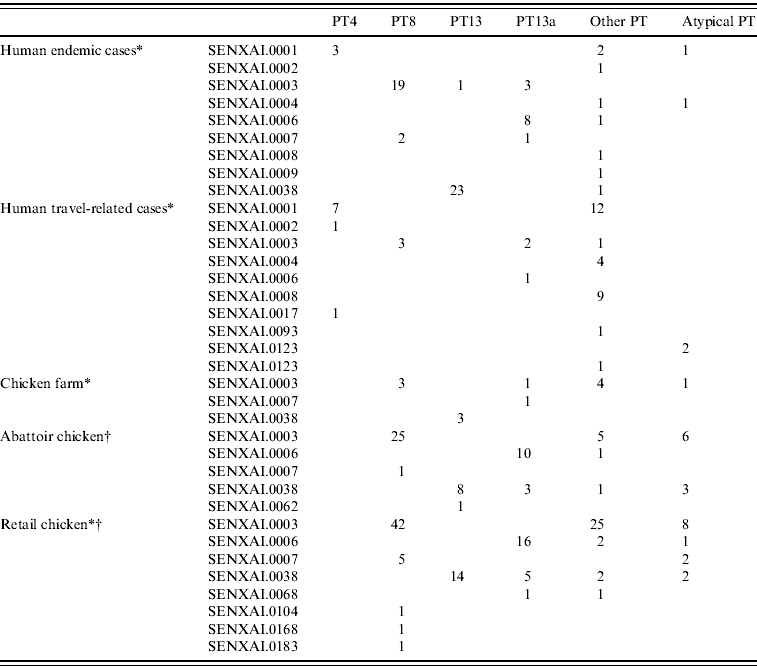
* C-EnterNet data (2005–2009 ).
† CIPARS non-human data (2008 and partial 2009).
Through CIPARS, 5691 human S. Enteritidis isolates were tested for antimicrobial susceptibility during the study period. Eighty-two percent (4683/5691) of isolates were susceptible to all antimicrobials tested, while resistance to ⩾1 antimicrobials was observed in 17% of isolates, and resistance to ⩾5 antimicrobials was observed in 1%. Resistance to one or more antimicrobials was most prevalent in PT1 strains (38%), followed by PT4 (19%), PT29a (4%), and PT6a (6%). Resistance was observed most often to nalidixic acid (14%), tetracycline (3%), ampicillin (2%), and sulfisoxazole (2%). All strains were susceptible to amikacin, two strains were resistant to ceftriaxone and one strain was resistant to ciprofloxacin. The resistance to tetracycline in all S. Enteritidis isolates increased significantly in 2007 compared to 2003.
S. Enteritidis in agri-food sources
S. Enteritidis in broiler chicken
Through CIPARS and C-EnterNet, S. Enteritidis was isolated from chicken on farms, slaughtered chicken, and retail chicken (Table 4). There are no results for chicken eggs since they are not actively monitored by these programmes. CIPARS data show that the relative proportion of S. Enteritidis in all Salmonella isolates recovered from slaughtered chicken increased from 0% (0/126) to 19% (44/230) over the study period (Table 4). The proportion of S. Enteritidis in CIPARS retail chicken increased from 0% (0/54) to 20% (94/473) and this upward trend was observed across all participating provinces. PT8, PT13a, and PT13 were the most common phage types recovered from the chicken commodity (Fig. 6, Table 2). All S. Enteritidis isolates from retail and abattoir chicken were fully susceptible to the antimicrobials tested except for two isolates from slaughtered chicken: one resistant to tetracycline and one resistant to streptomycin. Of all chicken S. Enteritidis isolates tested by PFGE, the most common pattern was SENXAI.0003, followed by SENXAI.0038 and SENXAI.0006 (Table 3). Pattern SENXAI.0003 was most closely associated with PT8.
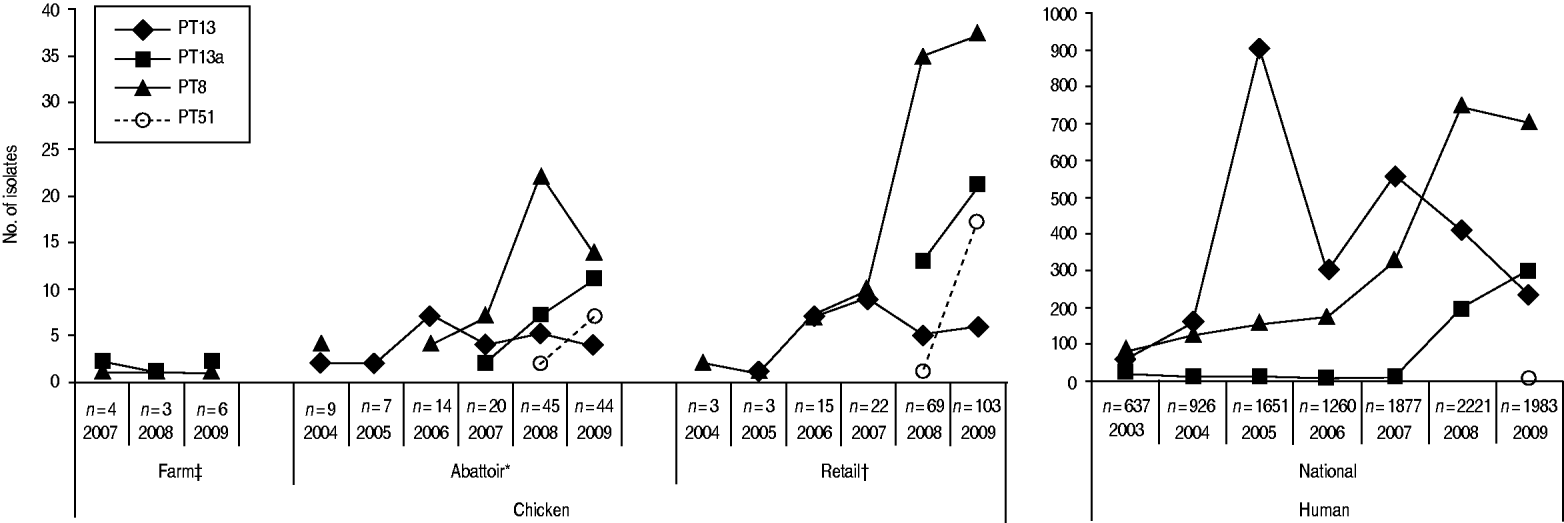
Fig. 6. Common S. Enteritidis phage types recovered in humans and non-humans, by year, in Canada, 2003–2009 [sources: National Microbiology Laboratory, Canadian Integrated Program for Antimicrobial Resistance Surveillance (CIPARS) and C-EnterNet]. * CIPARS data only. † CIPARS and C-EnterNet data combined. ‡ C-EnterNet data only.
Table 4. Number of Salmonella and S. Enteritidis recovered from non-human samples by active surveillance systems in Canada, 2003–2009 (source: Canadian Integrated Program for Antimicrobial Resistance Surveillance (CIPARS) and C-EnterNet)
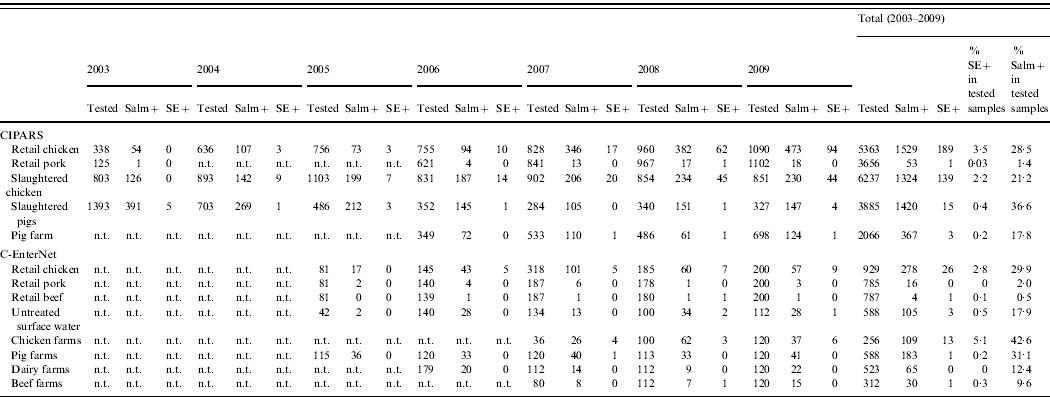
n.t., Samples not tested.
Salm+, Salmonella-positive isolates; SE+, Salmonella Enteritidis-positive isolates.
S. Enteritidis in other food animal commodities and the environment
S. Enteritidis was detected in pigs on farm, slaughtered pigs (caecal contents), and retail pork far less frequently than in the chicken commodity (Table 4). S. Enteritidis represented less than 1% (20/2039) of all Salmonella isolated from pigs on farm, at slaughter and at retail. Overall, PT8 was the most common phage type recovered from the pig commodity (Table 2). All pig isolates tested were fully susceptible to the antimicrobials tested. Two S. Enteritidis isolates were recovered by C-EnterNet from the beef commodity, one in retail ground beef and another in beef farm manure. Both were PT8 (Tables 2 and 4). Two S. Enteritidis isolates were detected in untreated surface water samples (Table 4), both were PT13.
S. Enteritidis in animal clinical isolates
Between 2003 and 2009, S. Enteritidis represented 11% (586/5261) of all Salmonella isolates recovered from all animal diagnostic submissions sent to STL. S. Enteritidis represented 0·66% (4/608) and 22% (191/861) of all diagnostic Salmonella isolates in 2003 and 2009, respectively. S. Enteritidis was isolated most often from chicken (471/586, 80%) and PT8 was the most common phage type (260/449 isolates with phage-type data, 58%) in chicken diagnostic isolates (Table 2). PT8 and PT13 were most commonly detected in diagnostic isolates from pigs, cattle and other animals (Table 2). Ten diagnostic S. Enteritidis isolates demonstrated antimicrobial resistance: three from chicken, two from cattle, two from pigs, and three from unspecified bird species. Four isolates showed resistance to ⩾5 antimicrobials; these included one pig and three chicken isolates. All chicken isolates showed resistance to antimicrobials of very high importance to human medicine: three were resistant to ceftriaxone and ceftiofur and two were resistant to amoxicillin-clavulanic acid.
S. Enteritidis from government monitoring programmes
S. Enteritidis was isolated from 15% (1188/7852) of all Salmonella isolates from government monitoring programmes submitted to the STL. Of these isolates, 93% (1107/1188) originated from chickens (e.g. broilers, broiler breeders, layers, layer breeders), ducks, turkeys or other avian species. The most common phage types across all government monitoring isolates were PT8 (43%, 516/1188), PT13 (15%, 174/1188), and PT13a (9%, 102/1188).
DISCUSSION
Through the integration of data provided by Canadian surveillance programmes, this study confirms and documents an increasing trend in Salmonella Enteritidis in the Canadian population and several exposure sources. S. Enteritidis has become the most common serotype of human salmonellosis in Canada, surpassing S. Typhimurium and representing about 33% of all human Salmonella isolates reported in 2009. The rates of S. Enteritidis infections in humans have increased threefold from 2003 to 2009, with most provinces and territories affected. The increase has been primarily associated with three main phage types, 8, 13 and 13a. These phage types had summer seasonal peaks and were associated with cases of domestically acquired infections. Similar phage types have been reported in the USA [Reference Marcus12, Reference Voetsch13]. In contrast, European countries have reported PT4, PT1, PT8, PT14B and PT21 as the most commonly identified phage types [14].
The various agri-food data examined in this study showed that the broiler chicken commodity was more often contaminated with S. Enteritidis than other livestock commodities that were regularly sampled. PT8, PT13 and PT13a were the most common strains in broiler samples, similar to human cases. The data showed an overall increase in recovery of S. Enteriditis from broiler chicken between 2005 and 2009, which paralleled that observed in humans. For example, a concurrent increase in PT8 was observed early on in the reporting period in slaughtered chicken, retail chicken, and humans, while an increase in PT13a was observed in slaughtered chicken, retail chicken, and humans in 2008 and 2009. Outbreak investigations involving PT8 and PT13 have implicated chicken and eggs as the source of human infection. These results are supported by the fact that broiler meat has recently been identified as a risk factor for S. Enteritidis in North America [Reference Voetsch13, Reference Kimura15] and that S. Enteritidis has been increasingly recovered from retail chicken meat [16–Reference Ribeiro18] and eggs [Reference Suresh19, 20]. In the USA, data (2001–2005) showed an increase in the proportion of PT8 isolates from chicken meat and a slight drop in the proportion of PT13 [Reference Altekruse21]. It is understood that gradual changes in predominant strains occur naturally over time; however, the exchange of broiler hatching eggs, chicks, and chicken meat products both inter-provincially [22] and internationally [23], may also contribute to the distribution and emergence of certain S. Enteritidis phage-type strains in Canada. Further investigation is needed to establish if changes in predominant phage-type strains occurring in both Canada and the USA occurred independently. Although monitoring of S. Enteritidis in this commodity is covered both by CIPARS and C-EnterNet, there is no current programme monitoring S. Enteritidis in imported poultry products to explain the increasing trend of S. Enteritidis and emergence of certain phage types in broiler chickens.
The egg commodity has also been recognized as a major S. Enteritidis exposure source for humans [Reference Marcus12, Reference Doorduyn24–Reference Braden28]. S. Enteritidis in this commodity is not monitored by either CIPARS or C-EnterNet. However, monitoring and intervention programmes are conducted by the egg industry and the Canadian Food Inspection Agency (CFIA) to ensure egg safety [29–31]. While some of these data are captured by government monitoring described above, the data available for this study were insufficient to determine which isolates were recovered from the egg industry vs. other poultry industries such as the layer industry. Currently, Health Canada is finalizing a policy on S. Enteritidis in Canadian shell eggs, which is informed by the risk assessment of Canadian Grade A shell eggs internally contaminated with S. Enteritidis [Reference DeWinter32]. This policy is intended to harmonize current inter-provincial differences in sampling and S. Enteritidis laboratory methods. This policy will enhance safety of eggs reaching the consumer by implementing early risk mitigation approaches in S. Enteritidis-positive laying flocks.
Beyond the egg and broiler commodities, S. Enteritidis was detected in other food animals (e.g. beef cattle, pigs), pets, wild animals, food (e.g. pork, beef, cheese, produce), and surface water, highlighting each as a potential source of human infection. However, few S. Enteritidis isolates have been recovered from these sources and the available data were insufficient to explore their contribution to the level and trend of S. Enteritidis infections in humans.
Antimicrobial resistance levels in S. Enteritidis sourced from healthy animals on farms and at slaughter, and from retail meats remained low over the study period. This is consistent with data from other surveillance programmes [16, 33]. In human isolates, resistance to nalidixic acid, tetracycline or ampicillin was observed in phage types that were not identified in food animal isolates (PT1, PT4, PT6a, PT29a). PT1 has repeatedly demonstrated the highest level of resistance to nalidixic acid and reduced susceptibility to ciprofloxacin [Reference Browning34–Reference Carrique-Mas36]. PT6a has also shown resistance to nalidixic acid [Reference Carrique-Mas36]. Currently, antimicrobial resistance in S. Enteritidis remains low. However, changes in resistance profiles should be monitored, especially for drugs of high importance to human medicine [37] such as fluoroquinolones and newer generation cephalosporins.
This study provides further insight into the epidemiology of S. Enteritidis infections in the Canadian population and the role of international travel. At the sentinel site level, international travel represents an important source of S. Enteritidis infection. Common phage types in travel-related cases included PT4, PT1 and PT6a. These findings corroborate with other studies, which identified PT4 [Reference Marcus12, Reference Voetsch13] and PT1 [Reference Molbak and Neimann38] as being associated with international travel. PT1, PT4 and PT6a are different from those contributing to the increase in endemic cases and are absent from all non-human sources in this study and from previous reports [Reference Poppe39–Reference Poppe41]. Of the CIPARS data, these phage types exhibited higher levels of resistance to nalidixic acid compared to all other phage types. In addition, these phage types showed a different seasonal pattern than the phage types associated with endemic cases, with larger peaks in the winter. While, S. Enteritidis infections acquired abroad appear to be different from infections acquired domestically, they do contribute to the overall burden of S. Enteritidis cases in Canada. This warrants enhanced surveillance and prevention efforts.
This study synthesized the best available information provided by many surveillance and monitoring programmes. Although each system has its own limitations, these synthesized findings provide a comprehensive picture of the epidemiology of S. Enteritidis in Canada. These results demonstrate the importance of ongoing national surveillance at different points in food production and the ability of integrated surveillance to identify human and animal health issues. Considering the increasing trend of human S. Enteritidis incidence in Canada and the difficulties in establishing epidemiological linkages between contaminated source and human disease, surveillance of this pathogen in humans, animals and the environment should continue and be conducted in a coordinated and collaborative manner. Future research should also be undertaken to elucidate the role of various potential sources in order to target public health actions to reduce exposure and to better understand the exposures and routes of transmission for human cases. Finally, enhanced collaboration between public health, animal health and the poultry industry is needed to implement effective prevention and control measures and mitigate the burden of illness caused by S. Enteritidis.
ACKNOWLEDGEMENTS
The authors acknowledge provincial and territorial epidemiologists and laboratory directors and programme stakeholders for their valued input on the manuscript. We also acknowledge provincial epidemiologists from the provinces of Ontario, British Columbia, Québec, and Alberta for providing additional outbreak information. The authors thank Andrea Ellis and Diane MacDonald for their advice and early discussions on the scope of the paper, Pia Muchaal and Lisa Scott for their assistance in the data analysis, Rebecca Irwin, Rafiq Ahmed, Frank Pollari, and Lisa Landry for their critical review of the manuscript. The authors especially thank the contribution provided by all the Provincial Public Health Laboratories for submitting isolates for further characterization and antimicrobial susceptibility testing. The Canadian Integrated Program for Antimicrobial Resistance Surveillance Public Health Partnership and the Canadian Public Health Laboratory Network including the provincial public health laboratories or enteric reference centres of British Columbia, Alberta, Saskatchewan, Manitoba, Ontario, Québec, New Brunswick, Nova Scotia, Prince Edward Island and Newfoundland and Labrador; the Laboratory for Foodborne Zoonoses, Guelph, Ontario and Saint-Hyacinthe, Québec; the National Microbiology Laboratory, Winnipeg, Manitoba; and the Centre for Food-borne, Environmental and Zoonotic Infectious Diseases, Guelph, Ontario.
DECLARATION OF INTEREST
None.












