INTRODUCTION
Acute otitis media (AOM) is a very common childhood disease that is responsible for many paediatric office visits [1]. Previous studies have shown that by the age of 3 years 80% of children will have experienced one episode of AOM, and by the age of 7 years 40% of children will have experienced ⩾6 episodes [Reference Casselbrant, Mandel, Rosenfeld and Bluestone2]. The peak incidence of AOM occurs between the ages of 6 and 18 months [Reference Teele, Klein and Rosner3]. AOM is associated with upper respiratory viral infections [Reference Vergison4–Reference Heikkinen and Chonmaitree10]. Children that suffer from recurrent or chronic episodes of AOM may experience temporary or permanent hearing loss, delayed language development, poor auditory processing, delayed psychosocial and cognitive development, and an adverse impact on educational progress [Reference Acuin11, Reference Monasta12].
Due to the large public health burden of AOM in children, in October 2000, a 7-valent pneumococcal conjugate vaccine (PCV7) was introduced into the routine schedule of immunizations for children [13]. At the time, seven serotypes (4, 6B, 9 V, 14, 18C, 19F, 23F) accounted for 80% of pneumococcal AOM in young children; that data dictated the serotypes included in PCV7 [Reference Pai14]. Shortly after the introduction of PCV7, there was a marked reduction in AOM caused by serotypes included in the vaccine [Reference Pichichero and Casey15–22].
In 2006, we began a longitudinal, multi-year, prospective study aiming to: (1) prospectively collect and study nasopharyngeal (NP) samples, middle ear fluid (MEF), and serum from young children (aged 6–30 months) who become otitis prone, who have infrequent otitis, or who remain otitis free; and (2) evaluate the role of specific immune responses to candidate vaccine bacterial proteins expressed by S. pneumoniae and non-typable H. influenzae (NTHi) otopathogens. We have previously reported on microbiological aspects of the project and on the immunological results. Our study population is the only group of children in the USA currently undergoing comprehensive, ongoing monitoring for potential risk factors for NP colonization and AOM, using tympanocentesis to prove every case of AOM. We sought to understand, risk factors that may influence changes in frequency of otopathogen infections as new vaccines are introduced. Here we used a relational database and statistical modelling to assess risk factors for pneumococcal NP colonization and AOM during the PCV7 era, 2006–2011.
DESIGN AND METHODS
Study community
Study participants were recruited from private paediatric offices in Rochester, NY. Rochester is located in Monroe County in Western NY. The population of Monroe County was 744 434 in 2010 (US Census Bureau: State and County Quick Facts), with <6% of the population aged <5 years. In Monroe County, 78·0% of the population identifies as White, 16·0% African American, 3·4% Asian, and 2·6% other. Of the 78% identifying as White, 72·6% identify as non-Hispanic White. The median household income for Monroe County in 2010 was $51303 and the total number of births for the year was 8466.
Child subject recruitment
About 85% of the subjects were recruited from a single private paediatric practice (Legacy Pediatrics, Rochester NY, main contributor, two paediatricians). Four other private paediatric groups joined in the recruitment effort by referral of patients to Legacy Pediatrics.
Eligibility
Children had to be aged at least 6 months to participate in this study. Inclusion criteria were: healthy, full-term birth, no craniofacial anomalies, no known immune deficits, and no AOM events prior to enrolment at age 6 months. All participants received four doses of PCV7 at ages 2, 4, 6, and 15 months until October 2010 when PCV13 was used to complete the PCV series depending on the date of their enrolment. Parental (both parents) consent was obtained prior to any study procedures. PCV7 contained the seven most prevalent serotypes causing invasive pneumococcal disease in young children at that time (4, 6B, 9V, 14, 18C, 19F, 23F). PCV13 contains 13 serotypes of pneumococci (serotypes in PCV7 plus 1, 3, 5, 6A, 7F, 19A) [Reference Shapiro23]. This study was approved by the University of Rochester IRB and subsequently by the Rochester General Hospital IRB.
Sampling
At the first visit, parents were asked to complete a questionnaire regarding demographic parameters and risk factors for AOM including: date of birth, gender/sex, race/ethnicity, height, weight, number and age of siblings, daycare attendance (none, centre, or home setting), breastfeeding status, exposure to tobacco smoke (defined as any smoker in the home), family history of OM, and allergies/atopy. Parents agreed to seven scheduled visits at ages 6, 9, 12, 15, 18, 24 and 30–36 months. At each of the above visits, parents were asked if there were any changes to the participant's demographic/risk factor history and whether the child had a current or recent upper respiratory tract infection within 2 weeks of the study visit. At each visit, the following biological samples were collected from the participants: nasal wash (NW), NP swab, blood, and throat culture (TC). At an unscheduled AOM visit, the following samples were collected: MEF (both or a single tympanocentesis procedure depending on whether the infection was bilateral or unilateral), NW, NP swab, blood, and TC.
Sampling procedures
For NW samples, 2 ml saline solution was squirted into the participant's nose with a rubber bulb syringe (Bard ear syringe). For NP samples, a wire nasopharyngeal flox brush was passed to the posterior nose and swabbed (Floq Swabs, Italy). For TC samples, sterile cotton-tipped applicators were used to swab the back of the child's throat (Select Medical Products, USA). For blood sampling, a venepuncture was performed and collection was into a heparinized tube. For MEF samples, an 8% tetracaine solution was instilled into the external auditory canal after placement of an otowick, then after ~15 min a 20-gauge 3·5″ spinal needle attached to a 3 cc syringe was used to puncture the tympanic membrane and collect MEF. All samples were collected at the paediatric office and transported to the laboratory via courier. For the remainder of this report, TC, NP, and NW samples will collectively be referred to as NP samples.
Microbiology and molecular biology
Microbiology processing, identification, and molecular testing for organism identification have been previously described [Reference Kaur24]. The four bacteria that were analysed in this report were Streptococcus pneumoniae, Haemophilus influenzae (non-typable), Moraxella catarrhalis, and alpha-haemolytic streptococci (AHS).
Digital data handling
All collected data were stored and accessed via a custom-developed AOM database application that consisted of a web-based user interface, data access and manipulation software, and a MySQL database. The application was designed with the following goals: to provide a reliable and accessible repository of project data, to provide screening tools for initial analysis of the data, to maintain the data and access to the data in a secure manner, to maximize extensibility, and to provide an intuitive user-friendly interface. Due to the breadth of the study objectives, samples from one patient's visit (i.e. a patient's third visit) could be used for multiple analyses; including otopathogen colonization in multiple samples (NW, TC, and in the case of AOM, MEF samples), antibody measurements (>1 antibody per otopathogen), and patient immunology responses; the in-house relational database was created to link all of these individual test results to the originating sample and patient. The database system was constructed to be compliant with the Code of Federal Regulations, Title 21, Part 11 that states all electronic records must be trustworthy and reliable [25]. This was achieved via a set of security and data integrity processes that were incorporated in the system. Data integrity was achieved by a combination of utilizing a fully normalized database schema to eliminate any data anomalies and a tightly controlled data input process that maximized user selection of predetermined values and mitigated erroneous data entry.
Statistical analyses
For the modelling analysis, the main outcomes of interest were the risk of AOM (AOM visits vs. non-AOM visits) and the risk of S. pneumoniae colonization in the NP of children (presence in the NP vs. no presence in the NP). All statistical analyses were conducted by using Stata version 12.0 (StataCorp., USA). Repeated-measures logistic regression with an unstructured correlation structure using the command xtmelogit was used for the analysis of the two models (the unit of study was the samples collected during a paediatric office visit). For both models, a univariate analysis was conducted first (a significance level of P = 0·10 was used for the univariate analysis), then all variables that were significant in the univariate analysis, were included in the full model. A backwards stepwise procedure was used to find the most parsimonious final model. A likelihood-ratio test was used to compare models as variables were dropped from previous to current models. If the two models changed significantly with the removal of a variable, then that variable was kept in the full model. Host factors that were included in this analysis were breastfeeding (<6 months vs. ⩾6 months), daycare attendance (centre/home setting vs. none), age (months), family history of OM (yes/no), siblings in the home (<5 years old, ⩾5 years old, both <5 and ⩾5 years old), sex, exposure to tobacco smoke (yes/no), and history of AOM episodes (⩾1) (yes/no). Results for the model were expressed as odd ratios (ORs) with 95% confidence intervals (CIs).
RESULTS
Recruitment results and demographics
During the 5-year period (June 2006–June 2011), 464 children were enrolled in the study. There were 5301 NP samples and 570 MEF samples collected (Table 1). In total, 159 patients completed the study by 30 June 2011, the end of the study period of this paper. Of the 159 patients, 21·4% completed all seven visits, another 31·4% completed 6/7 visits and the remaining 47·2% completed 5/7 visits. At the end of the 5-year time-frame for this report, the remaining 305 children enrolled were still participating in the study, and therefore had only completed a portion of their scheduled seven visits. Table 2 shows the demographic data for the study cohort. About 40% of the 464 patients experienced ⩽2 AOM events during their participation in the study up to 30 June 2011.
Table 1. Enrolment during the 5-year study (n = 464 patients with 1825 visits)
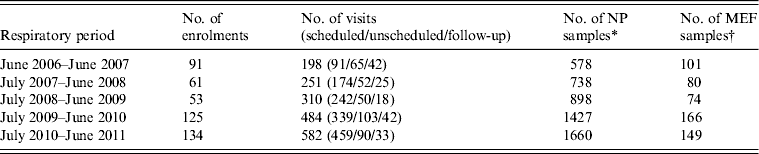
NP, Nasopharyngeal; MEF, Middle ear fluid.
* NP samples include throat culture, nasal wash, and NP swab.
† MEF samples include both unilateral and bilateral results in addition to broths collected for each MEF sample.
Table 2. Demographics of children enrolled during the study period (n = 464)

AOM, Acute otitis media.
* Missing data by variable: smoking status (n = 13); family history (n = 14); daycare (n = 8); sibling status (n = 7).
† There were 76 patients missing breastfeeding data, questionnaire captures data at the time of the visit.
Colonization
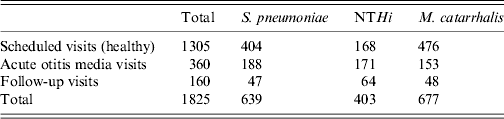
During the 5-year study there were a total of 1825 visits; 1305 (71·5%) were scheduled healthy visits, 360 (19·8%) were during episodes of AOM, and 160 (8·8%) were follow-up visits (3 weeks after an AOM episode) (Table 3). The predominant otopathogen during episodes of AOM was S. pneumoniae followed by NTHi. The predominant otopathogen during health visits was M. catarrhalis followed closely by S. pneumoniae. Compliance with a follow-up visit after an AOM episode was 44·3%; the predominant otopathogen in the NP at the follow-up visit was NTHi.
Table 3. Children enrolled during the 5-year study period (n = 1825 visits for 464 patients)
We observed 31 different serotypes of S. pneumoniae during the 5 years of this study. The most common serotypes/serogroups cultured during health visits were 19A (89 isolates), 15 (66 isolates), and 23B (40 isolates) and the most common during AOM visits were 19A (39 isolates), 15 (23 isolates), and 11 (nine isolates) in NP samples and 19A (25 isolates), 15 (seven isolates), and 11 (three isolates) in MEF samples (Fig. 1).

Fig. 1. Number of S. pneumoniae isolates by study period and circulating serotypes/serogroups. (a) Nasopharyngeal samples (total n = 548), (b) middle ear fluid samples (total n = 59). Not all circulating serotypes/serogroups are represented in this figure, as some were only present in <3 samples.
During the 5-year study, there were 360 visits due to AOM. A Pearson's correlation analysis of bacteria present in both the NP and MEF during these visits indicated that NTHi had the strongest positive correlation (0·44), S. pneumoniae had a weak positive correlation (0·25), and M. catarrhalis had no correlation (0·19) between presence of the bacteria in the NP and MEF during episodes of AOM.
Multilevel modelling
Repeated-measures logistic regression models predicting the risk of AOM by any otopathogen and the risk of S. pneumoniae colonization in the NP are shown in Table 4. In the model for risk of AOM, the following factors increased the risk of AOM: family history of OM (P = 0·02), NTHi in the NP (P< 0·01), S. pneumoniae in the NP (P< 0·01) and M. catarrhalis in the NP (P< 0·01). The presence of AHS in the NP decreased the risk of AOM (P < 0·01). In this model, breastfeeding for ⩾6 months increased the odds of AOM unexpectedly, but the result was not statistically significant.
Table 4. Predicted outcome of model A (AOM in young children) and model B (S. pneumoniae NP colonization in young children)

AOM, Acute otitis media; OR, odds ratio; CI, confidence interval; NTHi, non-typable Haemophilus influenzae; NP, nasopharynx; AHS, alpha-haemolytic streptococci; Ref., reference group; n.a., not applicable.
* Bold odds ratios indicate P value ⩽0·05.
† For random effects: estimate and 95% CI are given as well as the standard error.
In the second model (model B), the following factors increased the risk of S. pneumoniae colonization in the NP: children who attended daycare (P< 0·01) and who had M. catarrhalis present in the NP (P = 0·01). Children with siblings aged <5 years had an increased risk of S. pneumoniae NP colonization compared to children with no siblings (P = 0·01). In this model, children with a history of ⩾1 AOM episodes had an increased risk of S. pneumoniae colonization in the NP (P < 0·01). The correlation between S. pneumoniae colonization in the NP and history of AOM episodes was weak at 0·17.
We tested for linear age dependence by splitting the children into three age groups: <9 months, 9–18 months, and >18 months. Using the youngest group as the reference group in modelling we found age was not a significant factor in the models.
DISCUSSION
Here we report for the first time the results of the first 5 years of our ongoing prospective study on pneumococcal NP colonization and middle ear infections in children to the end of the PCV7 era in the USA. Results from this analysis show that S. pneumoniae was the most prevalent bacteria isolated from the NP during episodes of AOM and M. catarrhalis was the most prevalent bacteria isolated from the NP in healthy samples. The most prevalent serotype of S. pneumoniae isolates was 19A, in agreement with our earlier smaller study [Reference Casey, Adlowitz and Pichichero26] and others [Reference Pelton27, Reference Hanage28]. A new finding is the emergence of serogroups 15 and 11 as increasingly common NP colonizers and causes of AOM.
In our model, children with a family history of OM had an increased risk of AOM (OR 1·59, P = 0·02). A similar result was observed in families in Greenland [Reference Homoe29] and researchers have suggested a genetic link for OM risk [Reference Casselbrant and Mandel30]. In our study population we found that breastfeeding for ⩾6 months did not diminish the risk for AOM more than breastfeeding for a shorter time. We have no explanation for the non-significant trend of more frequent AOM in breastfed infants. In our study population we have previously reported a protective effect of breastfeeding [Reference Sabirov31].
Our results from the model for AOM confirm previous reports regarding the increased risk of AOM in children with S. pneumoniae, M. catarrhalis, and NTHi colonization in the NP [Reference Ruohola32, Reference Pettigrew33]. The model also showed that AHS colonization in the NP decreased the risk of AOM. This result is in agreement with our previous study [Reference Friedel34] and others [Reference Tano35] that upper respiratory commensals like AHS interfere with the colonization of potential otopathogens.
Host factors that increased the risk of S. pneumoniae colonization in the NP included daycare attendance and having siblings aged <5 years. Children who attend daycare and have siblings aged <5 years most likely attend daycare with their siblings; therefore, both children are exposed to outside pathogens. Daycare attendance is a known risk factor for AOM [Reference Cohen20, Reference Millar36].
The risk of S. pneumoniae NP colonization increased during concurrent M. catarrhalis NP colonization. Previous studies have observed a similar positive association between S. pneumoniae and M. catarrhalis [Reference Jacoby37]. Our group has found a synergistic (positive) association between M. catarrhalis and S. pneumoniae NP colonization in healthy children [Reference Xu38]. We have also observed a negative association between NTHi and S. pneumoniae and M. catarrhalis and NTHi NP colonization during episodes of AOM [Reference Xu38]. Our models indicated that a history of AOM episodes increases the risk of S. pneumoniae NP colonization in children. There may be genetic or epigenetic factors that influence the innate and/or adaptive immune response in the NP environment caused by intercurrent viral upper respiratory bacteria that facilitates the occurrence of AOM. Our group is now analysing the changing NP environment before and after AOM in otitis-prone children.
In late 2010, PCV13 became available in the USA ending the PCV7 era. The study period of June 2010–June 2011 showed a small reduction in serotype 19A prevalence in NP samples following the introduction of PCV13. With the anticipated reduction in serotype 19A, we predict other serogroups not included in PCV13 will fill this niche, similar to the serotype replacement observed after the introduction of PCV7 [Reference Wroe39].
This study is not without limitations. The study occurred predominantly in one, suburban paediatric practice site, although it is the only site in the USA conducting such prospective analyses of NP colonization and AOM events. Due to the voluntary basis of this study, not all patients were observed at all-time points (visits). The majority of children enrolled in the study who did not complete all seven visits were older than 6 months when enrolled and some children moved out of the Rochester area, therefore sample collection for these children was not complete.
CONCLUSION
This prospective, longitudinal study of a large cohort of children over the 5-year time-span 2006–2011 provides insight into the epidemiological risk and protective factors for NP colonization and AOM by pneumococci in children during the second half of the PCV7 era.
The study is unique due to the use of tympanocentesis to definitively prove every AOM infection. There were no other identical or similar studies of this nature ongoing in the USA or in the world to our knowledge so these are the only data collected for the time-frame of this investigation. The results reported here are highly relevant to those countries still exclusively or predominantly using PCV7. As we continue this study during the next 5 years in the same community, recruiting from the same population, using the same methods we will be able to compare the results reported here as we move into the PCV13 era.
ACKNOWLEDGEMENTS
This study was supported by NIH NIDCD RO1 08671. We thank the nurses and staff of Legacy Pediatrics and the collaborating paediatricians from Sunrise Pediatrics, Westfall Pediatrics, Lewis Pediatrics and Long Pond Pediatrics, the parents who consented and the children who participated in this study. We also thank Katerina Czup for her assistance.
DECLARATION OF INTEREST
None.







