Introduction
Although Q fever was first discovered in 1937, interest in the infection has re-awakened in recent years, partly because the potential for the causative agent to be used as a bioterrorism weapon and partly due to reported changes in the epidemiology in Europe [Reference Serbezov1–Reference Frankel3], including a number of large outbreaks [Reference Gyuranecz4–Reference Hawker7]. The largest of these recent outbreaks affected at least 4000 cases in the Netherlands from 2007 to 2010 [8].
Q fever is a zoonotic disease caused by Coxiella burnetii, a member of the γ-subdivision of Proteobacteria (Order Legionellales, Family Coxiellaceae). A number of animal species; sheep, goats, cattle, cats and dogs have been identified as sources of infection. Most infected animals are asymptomatic, although abortions may occur. In mammals, the infection localises to the endometrium and mammary glands and is reactivated during pregnancy to be aerosolised during parturition. These aerosols may be inhaled directly or may contaminate the environment for many months. The most common symptoms reported in acute infection are fever, headache, myalgia and cough. Other symptoms include fatigue, chills/rigors/night sweats, anorexia/weight loss, arthralgia and nausea/vomiting. The acute infection may occur as outbreaks. More rarely, chronic Q fever can follow clinically apparent or subclinical infection, possibly after a latent period of years and is estimated to occur in about 1–2% of cases [Reference Hawker9].
Knowledge of the incubation period of an infectious disease is vital in adequately responding to natural outbreaks or to a deliberate release of that agent. However, the quoted figures for the incubation period for acute Q fever vary substantially in the various Q fever review articles published in the literature. Estimates of the range vary from ‘a few days to several weeks’ [Reference Waag10], 4 days–6 weeks [Reference Parker, Barralet and Bell11], 10–17 days [Reference Raoult, Marrie and Mege12], 9–39 days [Reference Bossi13], 14–26 days [Reference Madariaga14], 1–3 weeks [Reference McQuiston and Childs15, Reference Maurin and Raoult16], 2–4 weeks [Reference Norlander17, Reference Aitken18], 14–39 days [Reference Fournier, Marrie and Raoult19, Reference Sawyer, Fishbein and McDade20] and 2–6 weeks [Reference Raoult and Marrie21]. Some state that incubation period is ‘usually’ 2–3 weeks [Reference Parker, Barralet and Bell11, Reference Bossi13, 22–24] or ‘approximately’ 20 days [Reference Fournier, Marrie and Raoult19]. Three reviews report an ‘average’, although this varies from 15 days [Reference Madariaga14] to 20 days [Reference Waag10, Reference Sawyer, Fishbein and McDade20]. One review [Reference Marrie and Raoult25] reports the results from early volunteer studies [Reference Tigertt and Benenson26] as a range between 10 and 16 days, and notes their finding that the incubation period was inversely related to the inhaled dose of the pathogen. The estimates of the incubation period given in nine of these reviews are unreferenced, five quote only other reviews/books and three give one or two papers with original data as references.
The value we can attach to the findings of systematic reviews intrinsically depends on both the way the review is conducted and the quality of the included studies. There are very few primary studies specifically designed to measure the incubation period of an organism; most incubation data come from reports of outbreaks of infection, which are usually focused on reporting the cause and/or management of the outbreak, with the incubation period usually being an incidental and often biased observation. Assessment of the robustness of the data on the incubation period provided in individual studies is vital to avoid contamination of the estimate by poor quality data (or if the best estimate is based on poor quality data, at least this can be acknowledged). Even in an exceptionally well conducted and reported outbreak study, there may be factors that make an analysis of the incubation period difficult to interpret. There are currently no tools or checklists to aid the assessment of the quality of the reported incubation periods. To address this in this paper, we used a conceptual, four domain framework to address the robustness of incubation period reporting. Here we reported a literature review summarising evidence of the incubation period of Q fever using a systematic literature search, a standardised assessment of the quality of data presented by each identified primary study and a synthesis of the results.
Methods
Study eligibility criteria
Studies reporting primary data on the incubation period of acute Q fever or data which allow incubation period to be calculated were eligible. Eligible designs were randomised control trials, non-randomised comparative studies, cohort studies (controlled or uncontrolled and including volunteer studies), outbreak reports, case studies and case series. Participants included males and females of any age with symptomatic, acute Q fever, and having experienced a defined exposure to Coxiella burnetti during a time period of less than one week. Studies not reporting incubation period information, where the course of disease may have been altered by chemoprophylaxis post-exposure (asymptomatic individuals are given antibiotics to prevent the development of symptoms following an exposure) or non-English publications were excluded.
Because many factors can influence the effectiveness of an intervention (e.g. patient characteristics, type of disease, variations in treatment, endpoints studied, etc.), systematic reviews of interventions usually define the study question with the aid of the four PICO (Patients, Intervention, Comparison, Outcome) elements [Reference Richardson27]. PICO has been shown to be a good framework for studies of interventions, but to be less suitable for studies of diagnosis, aetiology and prognosis [Reference Huang, Lin and Demner-Fushman28]. To address this, the PICO approach was modified by using the four elements defined in Box 1.
Box 1. ‘PICO’ approach to studying question design modified to address reviews of incubation periods
• Population studied: this may be the general population or a subset by, e.g., susceptibility (e.g. immunosuppression), age (e.g. children) or setting (e.g. hospital).
• Infectious agent: this may be all strains of the organism or a subset (e.g. influenza A and B may be analysed separately).
• Route or vehicle of infection: all routes of infection may be included or a subset (e.g. only waterborne or airborne exposure).
• Outcome: this will usually be the onset of the first symptom, but could be a specific symptom of public health interest (e.g. diarrhoea or coughing) or other markers of infection (e.g. seroconversion or excretion).
In terms of defining the review question: the population studied was all human cases; the infectious agent was C. burnetti; the route of infection was restricted to aerosol exposure (as we intended to replicate field conditions); and the outcome was the onset of symptoms consistent with a diagnosis of Q fever.
Search strategy
Electronic searches were performed conducted in Week 32, 2017 in Medline (OVIDSP) and EMBASE. No date restrictions were applied and search terms were left deliberately broad to ensure the search was sensitive enough to capture relevant studies. The English language only filter was selected. All identified abstracts from each of the databases were merged together in Refman v12, and duplicates removed using the ‘remove duplicates’ function. Reference lists of relevant systematic reviews identified in the searches were also checked, and suggestions by experts or conference findings included. Full details of the search strategy can be found in Supplementary material 1.
Study selection
Two reviewers (D.T. and T.F.) independently screened titles and abstracts of all identified records against the inclusion/exclusion criteria and subsequently independently reviewed full texts of all potentially eligible abstracts using the same criteria. Differences in opinion were discussed and agreed with the input of a third adjudicator (J.H.) where required.
Data extraction and management
Using a pre-piloted data sheet, two independent reviewers (D.T. and J.H.) extracted data as follows: study characteristics (author, the year in which the study was published, country, design and setting (e.g. rural, city, institution based)), outbreak information (case definition, likely source of infection, sample size, age group of cases and proportion of cases laboratory confirmed), incubation period information (was individual case data provided and/or was a median incubation period and range) and outcome measures (symptoms). Disagreements in extraction were reconciled by discussion between the reviewers.
If individual data were present or multiple outbreaks reported in one paper, data were selected which provided the best available evidence for incubation periods to form subsets, for example, if cases were laboratory confirmed or non-laboratory confirmed or the level of exposure differed amongst individuals; which might be considered at different levels of reporting robustness. Only those cases where a clearly measured incubation period could be determined were included. We also contacted the authors of some recent studies to obtain further information to help with the decision on whether to include the study and for quality assessments.
Assessment of robustness of incubation period reporting and role of bias
Studies reporting incubation period data and fitting the inclusion criteria were assessed independently by two reviewers (D.T. and J.H.). In evidence based medicine (EBM), assessing the quality of studies is usually carried out by using an existing checklist should an appropriate one exist, or developing one suitable for the question being studied if not. No such checklist currently exists for reviews of the data on incubation periods. Based on our experience of reviewing incubation data for other publications [Reference Waag10], we suggest four key criteria (each with three components) for assessing the quality of evidence provided in published reports giving data on incubation periods and summarise these in Box 2: this list is not organism-specific.
Box 2. Criteria for assessing the adequacy of individual studies reporting incubation periods of a pathogen
The Four Key Criteria for assessing individual studies are:
• Exposure – has the exposure to a source of the infectious agent been adequately demonstrated for the cluster and for individual cases?
• Diagnosis – has the outbreak been shown to be due to the organism and/or how reliable is the case-definition for individual cases?
• Accuracy – how reliable is the reporting of exposure and onset times?
• Ascertainment – are the inclusion and exclusion criteria for cases in which incubation reported appropriate for reducing contamination or bias?
Assessment of exposure includes:
• Is there a clearly defined exposure (e.g. attendance at a same function (e.g a farmer's market) and shared an exposure or eaten the same food)?
• Has this exposure been clearly linked to outcome, either epidemiologically or microbiologically?
• Have other potential exposures been adequately excluded?
Assessment of diagnosis includes:
• Has the cause of illness been microbiologically shown to be due to the study organism?
• If there is a clinical case-definition for some cases, is this sufficiently sensitive and specific?
• Are there time constraints in relation to onset/incubation in the case definition that limit the inclusion of very short or very long incubation periods?
Assessment of accuracy + precision of measures includes:
• Is there a clearly defined exposure time for cases?
• How reliable are the onset times, e.g. how was this collected and how long after?
• How accurately were incubation/onsets recorded, e.g. by hour/quarter day/whole day/week?
Assessment of risk of contamination or bias in included cases includes:
• Were all cases in the exposed group identified (e.g. cohort studies better than case-control or case series)? Is incubation/onset only reported on a subset of cases? Is this likely to introduce a bias?
• Contamination refers to if all cases within a sampling frame are truly all related to the exposure of interest, i.e. How well have background cases been excluded (this is particularly relevant during periods of high general community incidence)?
• If spread person to person*: How well have secondary cases been excluded (has there been subsequent mixing between some or all cases (e.g. at home, school or work?) and/or is there a risk that the shortest incubation case is, in fact, the source for the others (mainly relevant to the assessment of shortest incubation case)?
*Assumed to be uncommon for Q fever.
Papers were rated as either ‘good’, ‘moderate’, ‘low’ or ‘very low’ robustness of the incubation period data in the paper. We gave each study a score based on each of the four key criteria in Box 2: each criterion score started as zero and lost 1 point for a minor flaw, 2 points for a major flaw and 6 points for a ‘terminal’ flaw. The overall quality rating was then based on the scores being added across the four criteria, with a score of 0 equating to good evidence, −2 to moderate evidence, −4 to low evidence and −6 to very low. A ‘terminal’ flaw (a score of −6) in any of the criterion (e.g. failure to demonstrate that the outbreak is due to the organism of interest) automatically led to a rating of very low-quality evidence. As the majority of studies are not designed to either investigate the incubation period or necessarily report it accurately, it is important to highlight that the rating does not reflect the quality of the study; instead, it is a measure of the robustness in which the incubation period was reported during that individual study.
Data synthesis and analysis
Those studies which were considered to provide good or moderately robust evidence of the incubation period and we were able to access either individual data were included in the pooled analysis. For comparison, studies rated as ‘low’ and with individual-level data were also pooled. We also assessed whether the pooled data fitted a normal distribution by using the Shapiro–Wilk test, or could be transformed into a normal distribution (e.g. the log-normal distribution previously reported for many infectious diseases [Reference Sartwell29]) using the ‘ladder’ command in STATA V12. Normally transformed data would enable modelling which would allow the incorporation of data from studies that only gave summary data, (rather than individual data), into a modelled composite distribution.
Results
A total of 11 888 records were identified through the database search. Two articles [Reference Holland30, Reference Tigertt31] were found through hand searching of reference lists from identified articles and an additional study was identified at a conference (this has now been submitted for publication and a full draft kindly supplied to us by the authors) [Reference Wagner-Wiening32]. After duplicates were removed 7115 titles and abstracts were screened, with 6741 not meeting the inclusion/exclusion criteria and excluded at this stage. The majority of studies were rejected due to a lack of relevant data. Of the articles which required full paper review (n = 374), nine were non-English language, and we were unable to access 45.320 full articles or conference presentations were assessed for eligibility, 22 of which reported on unique outbreaks or cases, and two papers reported on the same study [Reference Tigertt and Benenson26, Reference Tigertt31] and are considered as one study in this review. The majority of the excluded studies (n = 351) were excluded due to insufficient information on incubation period (or it could not be derived) (n = 259), no clear exposure or exposure time period (n = 31) not primary data (n = 3) or non-aerosol route of acquisition (n = 3). The PRISMA [Reference Moher33] diagram is provided as supplementary material.
Robustness of incubation period reporting in identified studies
Based on our four domain assessment of the quality of the data (Exposure, Diagnosis, Accuracy/Precision and Contamination), two studies [Reference Tigertt31, Reference Dupont34] provided good quality estimates of the incubation period; seven papers [Reference Whelan5, Reference Wagner-Wiening32, Reference Kosatsky35–Reference Pinsky39] and a subset from an eighth paper [Reference Porten6] provided moderate quality estimations of the incubation period. Seven papers [Reference Langley40–Reference Selvaggi46] plus an additional subset from a paper also providing the moderate quality subset [Reference Porten6] were evaluated as providing low-quality data and six papers [Reference Holland30, Reference Harvey, Forbes and Marmion47–Reference Qazi51] as providing very low-quality data of the incubation period. A summary of the rationale behind the quality estimations for individual papers can be found in the supplementary material.
The two studies providing good quality data [Reference Tigertt31, Reference Dupont34] were volunteer studies with exposure and symptom monitoring done in a controlled environment. The papers providing moderate quality data were all real-world outbreaks (as opposed to volunteer studies), but with varying modes of transmission and in different geographical locations. Two of the papers described outbreaks from Canada [Reference Kosatsky35, Reference Buhariwalla, Cann and Marrie36], two from Germany [Reference Porten6, Reference Wagner-Wiening32] and others from the USA [Reference Buhariwalla, Cann and Marrie36], UK [Reference Marmion and Stoker37] and the Netherlands [Reference Whelan5]. The outbreaks vary in the source of infection; cats [Reference Kosatsky35, Reference Pinsky39], a dog [Reference Buhariwalla, Cann and Marrie36], an autopsy [Reference Marmion and Stoker37] and sheep [Reference Whelan5, Reference Porten6, Reference Wagner-Wiening32, Reference O'connor, Tribe and Givney38] were implicated. The identified studies and data identified from these studies are summarised in Table 1.
Table 1. Summary of identified studies
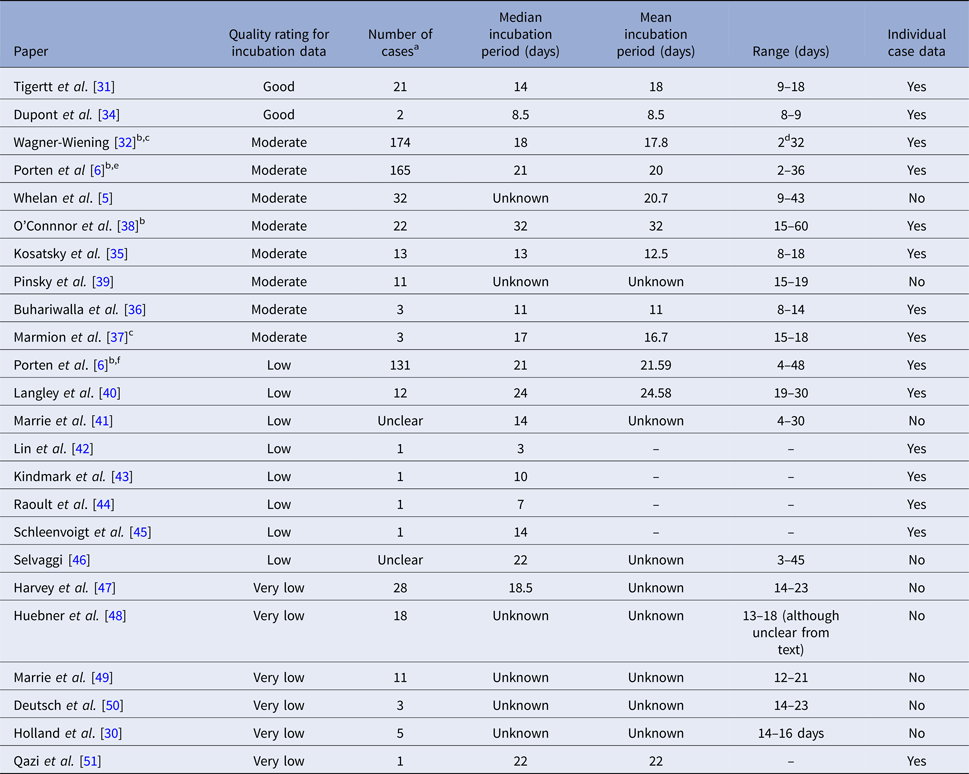
a Number of cases where the derivation of the incubation period was possible.
b Information from published study/poster supplemented through personal communication with authors.
c Selected subset of cases which provides information on incubation period.
d Differs from information provided on an epidemic curve, minimum incubation period taken from text.
e Laboratory confirmed cases only.
f Non-laboratory-confirmed cases.
Incubation period
The two studies rated as providing good quality information on incubation period provided individual-level data. We were able to extract individual-level information from six of the eight studies providing moderate quality data [Reference Porten6, Reference Wagner-Wiening32, Reference Kosatsky35–Reference O'connor, Tribe and Givney38] (n = 380), one of which [Reference Porten6] a subset was considered ‘moderate’ (laboratory-confirmed cases) and a further subset considered ‘low’-quality data (clinical cases). This subset and five other individual studies provided ‘low’-quality individual-level data (n = 146).
We could find good quality information on the incubation period for Q fever for 23 cases from two volunteer studies: these showed a range of incubations of 8–18 days, with a median of 14. There was clear evidence that the incubation period was inversely proportional to the inhaled dose in the larger [Reference Tigertt31] (21 cases) of the two studies.
Individual-level data could be extracted on 403 individuals from papers with good or moderate quality data (the two volunteer studies and six outbreak reports) and these were merged into a combined distribution in Figure 1: 95% of these cases fitted within an incubation period of 7–32 days (and 99% within 2–50 days), the median incubation was 18 days and the mean 19 days. There were two further outbreaks [Reference Whelan5, Reference Pinsky39] that were rated as providing moderate quality evidence for incubation period data, which reported the range and/or mean for the incubation period, but did not report individual-level data (or produce an output, such as an Epidemic Curve, from which we could derive it). Because Figure 1 did not demonstrate a log-normal distribution nor could be transformed to follow a normal distribution, we did not mathematically combine these additional data, using a log-normal (or any other) assumption, but the inclusion of another 43 cases with an approximate mean of 19·75 days (we assumed that the mean for Pinsky [Reference Pinsky39] was 17 days for this rough calculation) is similar to our observed mean of 19 days and it is unlikely inclusion would alter our above estimate of the median substantially in a new (theoretical) combined dataset of 446 cases.
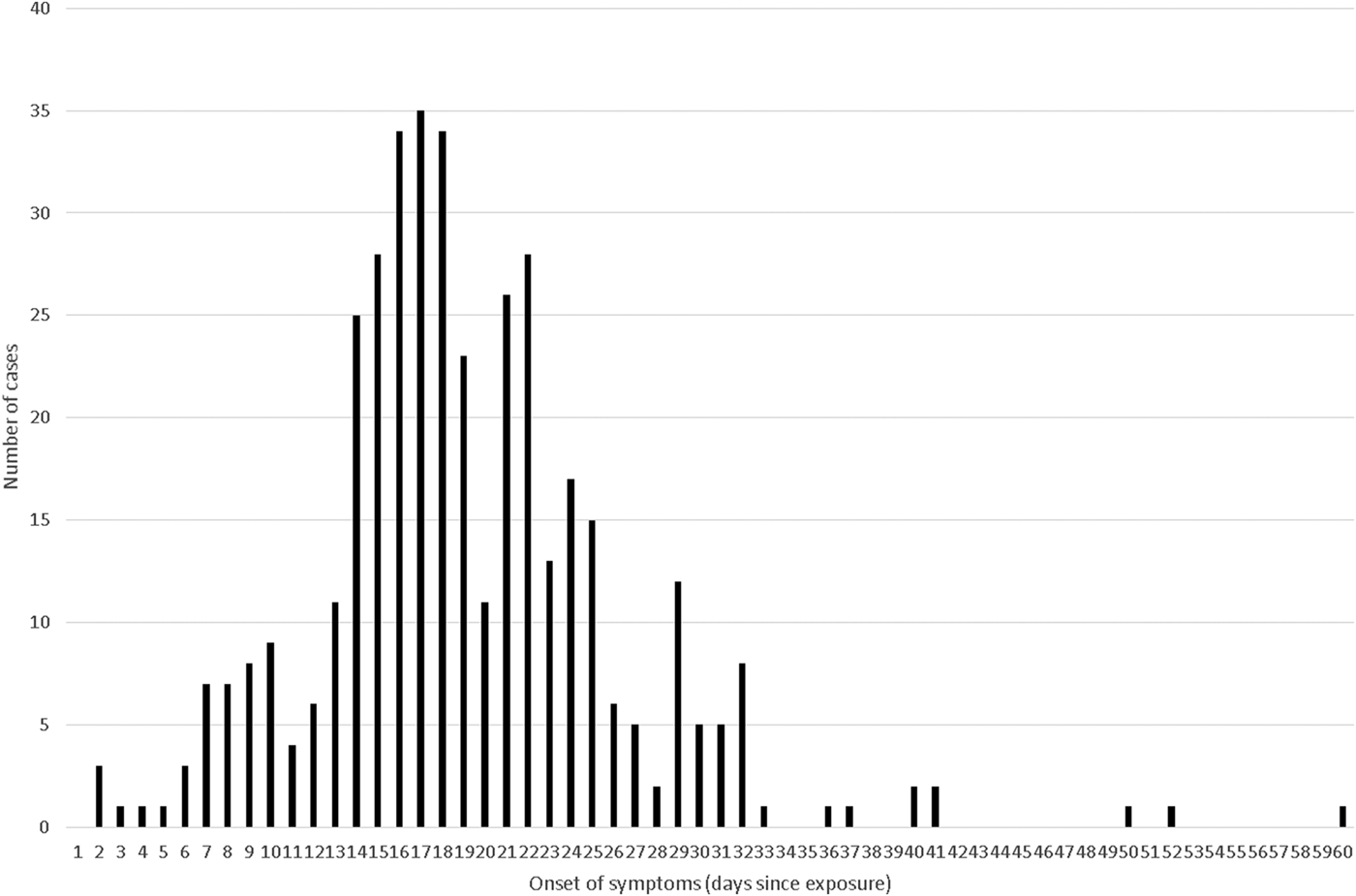
Fig. 1. Pooled incubation periods from studies rated ‘good’ or ‘moderate’ and with individual-level data (n = 403).
Adding the observations from five studies and a subset of a sixth that provided low quality rated individual level data to the distribution gave a combined curve covering 549 individual observations (Fig. 2). 95% of these observations fitted within an incubation period of 7–33 days (and 99% within 2–50 days), the median incubation was 19 days and the mean was 20 days.
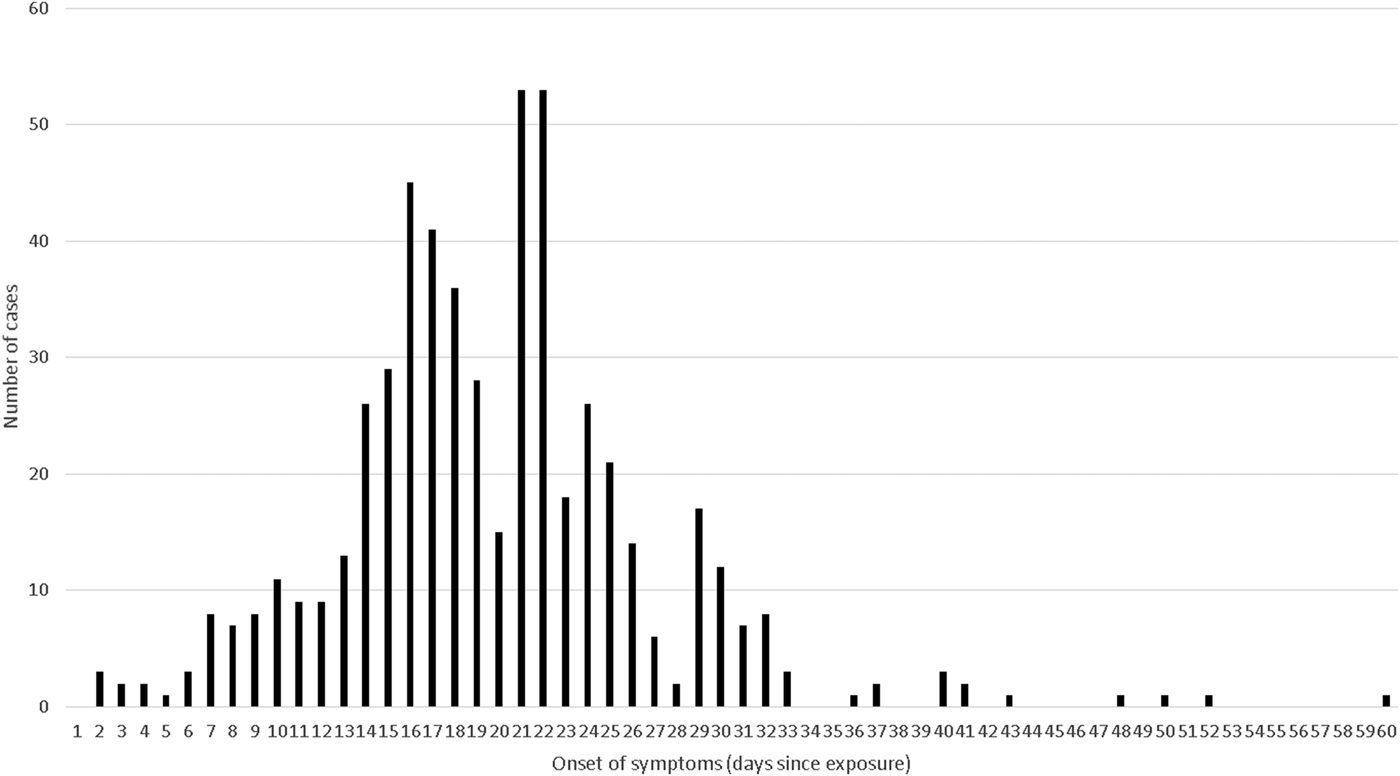
Fig. 2. Pooled incubation periods from studies rated ‘good’, ‘moderate’ and ‘low’ and with individual-level data (n = 549).
Discussion
There have been no previous systematic reviews of the published evidence for the incubation period for Q fever and the various information sources available to the public health and scientific community sources quoted in the introduction showed wide variation in the estimates given, with the minimum incubations varying from 4 to 14 days and maximum varying from 17 to 42 days (10–25).
The median incubation period of Q fever from studies which provided at least moderately robust reporting of individual-level data is 18 days. As 95% of observations from individual-level data of at least moderate quality data demonstrates an incubation period of between 7 and 32 days (and the inclusion of additional low-quality data made little difference to this result), we suggest that this should be reported as the ‘usual’ incubation period range.
There are reports of incubation periods of as low as 2 days, and as high as 60 days. However, the data on these outliers are not necessarily of the same quality as the rating of the individual study datasets that they are extracted from, e.g. some outbreaks had varying levels of background incidence that could contaminate the outbreak cases (a particular risk for outliers), and strain typing to eliminate background cases is not usually available. The 60-day incubation report appears to be a single outlier (Fig. 1) and may need to be treated with some caution. There were three separate laboratory-confirmed cases in contact with the demonstrated source from two separate papers with an incubation period of two days and two more cases each for 3 and 4 days, so this is less obviously an outlier and cannot be ruled out, particularly if the cases might have received a particularly high infecting dose or have some other reason to be particularly susceptible. There was good evidence from one of the experimental studies that incubation period for Q fever is dose dependent [Reference Tigertt and Benenson26, Reference Tigertt31], and a low infective dose was postulated as a possible reason for a higher than usual median incubation period in one of the identified outbreaks [Reference O'connor, Tribe and Givney38]. We suggest that the 2–50-day range covering 99% of cases in both or datasets is currently the best estimate for the extremes of the incubation period for Q fever.
Only 23 individual observations were found from studies that provided good quality data and these were from studies in which volunteers were exposed to an experimentally induced inhalation: it is not known how well these studies reproduce a natural exposure and so the validity of extrapolating this small number of experimental observations into an incubation period range for natural outbreaks is unclear. There is a need for further data from natural Q fever outbreaks, particularly from large outbreaks or from an outbreak of any size that can contribute data of good quality in terms of exposure definition, case definition, the accuracy of recording and lack of contamination.
Strengths and weaknesses
The main strength of this review was a systematic approach with an a priori determined study eligibility criteria (using an adapted PICO framework) and methodology that were applied to the research question formulation, selection, extraction, quality appraisal and synthesis of relevant evidence. Such a quality appraisal process is something that has not been systematically applied in the previously published studies of incubation periods for other infections that we were able to identify. The specific tool that we used provided quality scores in this study were felt by reviewers to accurately reflect their subjective view of the quality of the data extracted, and testing in further studies is underway.
One of the limitations of this review was the inability to assess publication bias which may have been present due to the exclusion a number of studies we were unable to retrieve and the exclusion of non-English language publications.
The main limitation of this review rests upon the evidence itself. Out of the numerous reports of Q fever outbreaks or cases, only a comparatively small number of studies or scenarios describe a temporally short (and single) well-documented exposure to a well-defined causative agent which allows determination of accurate incubation periods. Even fewer studies are designed to specifically measure incubation periods and it is often reported as an incidental finding, even in those studies where there is a likely single exposure to the causative organism. Incubation period information is usually used in outbreak investigations to link cases with probable – and often multiple – exposures [Reference Hawker7] which do not enable determination of the actual incubation period, but instead highlights the need for an accurate estimation.
An additional limitation is that the majority of cases were from two individual studies, one of which [Reference Porten6] itself demonstrated an unexpected bimodal distribution of data; with possible dose-dependent effect on incubation period. This both precluded a meta-analysis or statistical synthesis of the data, and may have masked the ‘true’ median incubation period. The second study [Reference Wagner-Wiening32] with a large amount of data was a poster presentation and a draft paper for publication that was yet to be peer-reviewed. Based on the extraction of the data from the epidemic curve, there appeared to be minor differences between our extracted data and the authors’ summary of the data, although these are unlikely to have affected our pooled estimates.
Conclusion
Based on the available literature which provides at least moderate robustness in the reporting of the incubation period, the median incubation period for Q fever is 18 days, with 95% of cases expected to occur between 7–32 days after exposure, although incubation periods between 2 and 50 days cannot be ruled out.
This review has highlighted the need for further good quality primary data on the incubation period for Q fever and for clear reporting of incubation periods in the reporting of outbreaks to build the evidence base for public health actions and decisions, which depend heavily upon accurate measures of epidemiological parameters.
Supplementary material
The supplementary material for this article can be found at https://doi.org/10.1017/S095026881700303X
Acknowledgements
We wish to thank the authors from the papers by Porten et al. [Reference Porten6] and O'Connor et al. [Reference O'connor, Tribe and Givney38] who kindly provided additional information or data. We wish to acknowledge Professor Anthony Stewart's contribution in developing the conceptual methodology for assessing the quality of incubation period reporting.
Conflict of interest
None declared.






