INTRODUCTION
Human parechoviruses (HPeVs) of the family picornaviridae are small, non-enveloped, positive sense, single-stranded (ss) RNA viruses. These viruses, previously known as Echo 22 and 23, were first isolated from the children during a summer outbreak of diarrhoea [Reference Wigand and Sabin1]. Subsequently, genomic analysis revealed them to be distinct from Echoviruses and classified into a new genus parechovirus [Reference Stanway and Hyypia2]. The HPeV genome is ~7·4 kb nucleotides in length and contains a large open reading frame coding for a single polyprotein which is post-translationally cleaved into three structural (VP0, VP3 and VP1) and seven non-structural (2A–2C and 3A–3D) proteins [Reference Stanway, Joki-Korpela and Hyypia3]. Based on the VP1 gene analysis, HPeVs have been classified into 17 genotypes i.e., HPeV1-17 (http://www.picornastudygroup.com/types/parechovirus/hpev.htm).
HPeVs are widely distributed and have been reported to cause various clinical manifestations including gastroenteritis, upper respiratory tract infections, sepsis-like illness, aseptic meningitis, Reye's syndrome, myocarditis, encephalitis and transient paralysis [Reference Baumgarte4]. Among HPeVs, HPeV1 is a widespread pathogen most commonly associated with gastrointestinal and respiratory symptoms and less frequently reported in severe disease conditions, encephalitis and flaccid paralysis [Reference Benschop5]. HPeV2 infections rarely occur and are associated with gastrointestinal symptoms [Reference Ehrnst and Eriksson6]. HPeV3 and HPeV4 were isolated from neonates with sepsis and also with high-grade fever and poor feeding, respectively [Reference Benschop5, Reference Ito7]. HPeV5 formerly classified as HPeV2 by neutralisation test has been reclassified on the basis of phylogenetic analysis of the VP1 region [Reference Oberste, Maher and Pallansch8]. HPeV6 was isolated from a child suffering from Reye's syndrome [Reference Watanabe9] while HPeV7 has been detected in a healthy child having close contact with a person who had acute flaccid paralysis [Reference Watanabe9, Reference Li10]. HPeV8 was isolated from a child with enteritis during an outbreak of diarrhoea in Brazil [Reference Drexler11]. HPeV9-13 were discovered in Thailand while HPeV14 was in the Netherlands. HPeV 15 and 16 have been reported recently from Bolivia [Reference Nix12].
Acute gastroenteritis (AGE) is a global health problem and a major contributor to childhood morbidity and mortality worldwide. Approximately two million deaths occur due to acute gastroenteritis in children under 5 years of age in the developed and developing countries [Reference Elliott13]. In India, an association of enteric viruses such as rotavirus (RV), norovirus (NoV), adenovirus (AdV), astrovirus (AstV) and other picornaviruses with acute gastroenteritis has been documented [Reference Kang14–Reference Verma, Chitambar and Gopalkrishna18]. Although variable rates (1·6–29·4%) of HPeV infections and their genotypic distribution in acute gastroenteritis cases have been reported worldwide [Reference Zhang19], no such reports are available from India. The present study was aimed to gain insight into the epidemiological features and genetic diversity of HPeV strains circulating in hospitalised children with acute gastroenteritis. This is the first report that describes the epidemiological and clinical features and genetic characterisation of HPeVs in acute gastroenteritis cases from India.
METHODS
Specimens
Stool specimens (n = 979) collected from children, aged <1–60 months (median age 12 months), hospitalised for acute gastroenteritis in Pune, western India during January 2006–December 2010 were included in the study. A sporadic case of acute gastroenteritis was defined as ⩾3 episodes of diarrhoea reported within 24 h with or without associated symptoms such as vomiting, fever and abdominal pain. One specimen per patient was collected for this study within 24 h of hospitalisation and prior written informed consent was taken from the parents/guardians. All the patients were examined for fever, number of episodes and duration of vomiting and diarrhoea, the extent of dehydration and treatment for the assessment of disease severity [Reference Ruuska and Vesikari20].
Nucleic acid extraction and detection of HPeV and other enteric viruses
Viral nucleic acid was extracted from 30% (weight/volume or volume/volume) of the stool suspended in 0·01 M phosphate buffered saline (PBS) pH 7·2 by using QIAamp viral RNA mini kit (Qiagen, Hilden, Germany) and stored at −80 °C until used. Presence of HPeV-RNA was identified by reverse transcription-polymerase chain reaction (RT-PCR) using 5′UTR specific primers followed by genotyping with primers specific for VP1 gene [Reference van der Sanden21, Reference Nix22].
The specimens were also tested for the presence of group A rotavirus antigen by ELISA (Dako Cytomation, Ely, Cambs, UK) and NoV, AdV, AstV and EV nucleic acids by RT-PCR/PCR as described earlier [Reference Chhabra15–Reference Verma, Chitambar and Gopalkrishna17, Reference Sapkal23].
All the PCR products were electrophoresed in 2% agarose gel containing ethidium bromide (0·5 µg/ml), visualised under UV transilluminator and products were sequenced with an ABI Prism 3700 DNA Analyser (Applied Biosystems, USA).
Nucleotide sequence analysis of HPeV genes
Sequence identity of VP1 gene of the HPeV strains was determined through BLAST analysis (http://www.ncbi.nlm.nih.gov/blast). Multiple sequence alignment was carried out using CLUSTAL W and phylogenetic analysis was carried out using MEGA 5.0 and the tree was generated with maximum likelihood method and Kimura 2-parameter distance model. The reliability of the phylogenetic tree was tested by applying the bootstrap test with 1000 bootstrap replications [Reference Tamura24]. The nucleotide sequences reported in this study have been deposited in GenBank under accession numbers KJ743617–KJ743718.
Statistical methods
The descriptive statistical analysis was done in Microsoft Excel 2007 while the prevalence between different groups was compared using χ 2 tests. All these tests were performed at the significance level of 0·05 using Open Epi 2.3.1 version and all those tests were two-tailed. Odds ratios (OR) and 95% confidence intervals (CIs) were also calculated.
RESULTS
Prevalence of HPeV among children with acute gastroenteritis
At least one enteric virus was detected in 602 (61·5%) of the 979 stool specimens collected from the children hospitalised for acute gastroenteritis in Pune, western India during 2006–2010.
Overall, RV, EV, NoV, AdV and AstV were detected in 242 (24·7%), 118 (12·1%) 65 (6·6%), 29 (2·9%) and 12 (1·2%) of the specimens respectively. HPeV was detected in 13·9% (136/979) of the specimens tested which comprises 56·6% (77/136) of HPeV mono infections and 43·4% (59/136) co-infection of HPeV with other enteric viruses. Among co-infections, dual infections were detected in 52 (88·13%) and triple infections in seven (11·86%) of the specimens. Rota virus (33·9%) and enterovirus (32·2%) were detected most frequently along with HPeV.
The overall prevalence of lone HPeV among acute gastroenteritis cases which are negative for other enteric viruses was observed to be 14·1% (77/548) (Table 1). The yearly prevalence of HPeV in stool specimens varied during the study period. HPeV was detected in 9·6% and 10·7% of the patients during 2006 and 2010, respectively and 6–8% in the remaining years (Fig. 1).
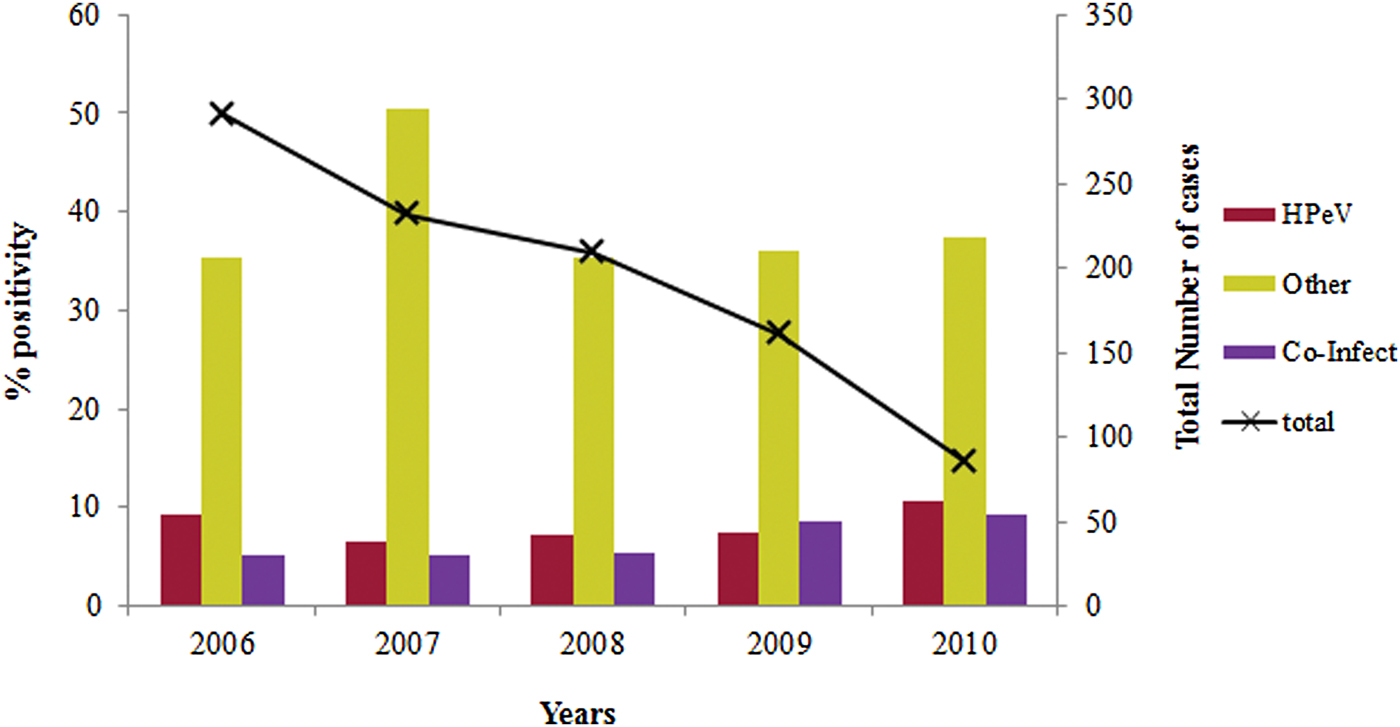
Fig. 1. Year wise distribution of HPeV in acute gastroenteritis cases during 2006–2010.
Table 1. Clinical and demographic features of human parechovirus (HPeV) infections among children with acute gastroenteritis

Ot Ent, other enteric viruses; Co-Inf, co-infection.
Epidemiological characteristics of HPeV infection among children with acute gastroenteritis
Age of the HPeV positive children (n = 136) was in the range 1–54 months (median, 10 months). The detection rate of HPeV decreased with the increase in their age and it was found more frequent in children ⩽1 year of age. Among children in ⩽1 year of age group, HPeV positivity was found significantly high (68·8%) as compared with the children tested positive for other enteric viral infections (χ 2 = 4·3; P < 0·05; OR = 1·8) and negative for all enteric viral infections (χ 2 = 8·6; P < 0·005; OR = 8·6) (Fig. 2).
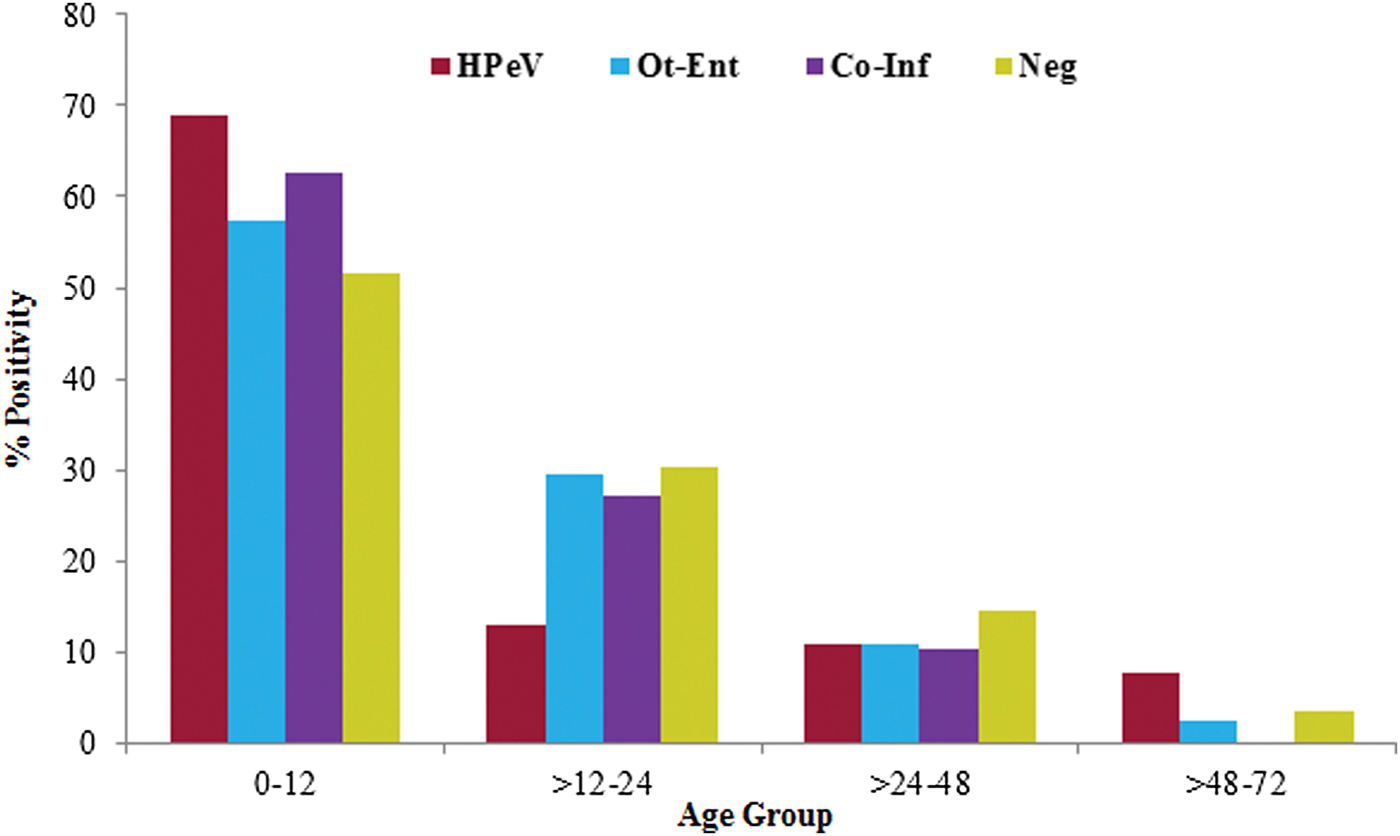
Fig. 2. The age distribution of HPeV positivity in acute gastroenteritis cases.
Among HPeV mono infected children, 27(35·5%) of them were females and 50(64·9%) were males. The female to male ratio for HPeV infection was 1 : 1·85 (Table 1). The HPeV incidence was documented throughout the year with a low prevalence (<10%) observed during the spring season (March and April).
Clinical features of HPeV infection among children with acute gastroenteritis
Among the HPeV infected children, 37·7%, 44·2% and 18·2% of the children showed clinical features such as no fever, low-grade fever and high-grade fever, respectively according to the vesikari clinical severity scoring method [Reference Ruuska and Vesikari20]. As compared with the children infected with other enteric viruses (χ 2 = 6·8; P < 0·05; OR = 2·6) and children without any viral infections investigated in this study (χ 2 = 5·2; P < 0·05; OR = 2·2), significantly more number of children with high- grade fever was recorded in HPeV infection (Table 1). Among the HPeV infected children, diarrhoea lasted for 1–12 days (Mean = 3·0 days) and the frequency of stool passage ranged from 3 to 25 times (Mean = 6·0 episodes) per day. Among the children infected with HPeV and other enteric viruses, diarrhoea lasted for 1–8 days (Mean = 3·1days) and the frequency of stool passage ranged from 3 to 15 times (Mean = 5·7 episodes) per day. Also, no significant difference was observed for different diarrheal condition between children infected with HPeV, other enteric viruses and children without virus infection (Table 1).
In the study, 48 (62·3%) children having HPeV mono infections and 40 (67·8%) children co-infected with HPeV and other enteric viruses experienced with vomiting. It was also observed that the vomiting lasted for 1–8 days (Mean = 2·2 days) and the frequency of vomiting episode ranged 2–8 (Mean = 3·4 episode) per day. The frequency of 1–4 episodes of vomiting in a day was significantly higher in HPeV infected children (χ 2 = 3·8; P < 0·05; OR = 1·6) and other enteric virus-infected children (χ 2 = 6·9; P < 0·05; OR = 1·4) as compared with the children tested negative for virus infection (Table 1). The disease condition based on Vesikari score was analysed and a significantly higher number of severe acute gastroenteritis cases was recorded in only HPeV infected children (n = 49; χ 2 = 4·6; P < 0·05; OR = 1·8) and among the children with co-infection of HPeV and other enteric viruses (n = 40; χ 2 = 6·2; P < 0·05; OR = 2·1) as compared with the children without virus infection (Table 1).
Genotyping and identification of HPeV strains in acute gastroenteritis
Out of the 136 specimens positive for HPeV-RNA, 102 (75%) were genotyped. Of these, 72 strains were successfully amplified and sequenced for full VP1 (880 bp) gene and the remaining 30 strains for partial VP1 (658 bp) gene. Phylogenetic analysis identified a total of 13 genotypes among the 102 strains, based on the criterion described earlier (22). The most prevalent genotypes observed were of HPeV1 (39, 38·2%), HPeV3 (13, 12·7%), HPeV4 (13, 12·7%) and HPeV5 (11, 10, 7%). Other nine HPeV types which included HPeV14 (4), HPeV2 (4), HPeV8 (4), HPeV10 (3), HPeV6 (3), HPeV13 (3), HPeV7 (2), HPeV16 (2) and HPeV11 (1) also were detected at lower level.
The strains identified as HPeV1 were grouped into two clusters i.e., HPeV1A (84·6%, 33/39) and HPeV1B (15·4%, 6/39). The nucleotide (NT) identity between the HPeV1A Indian strains and the prototype strain (Harris strain, 1956) was noted as 80·58–84·02%. The six strains in HPeV1B showed 90·35–92·37% NT identity with the strains from the Netherlands, AB112487 and FM178558 (Fig. 3).

Fig. 3. Phylogenetic dendrogram on the basis of VP1 gene of HPeV strains. Study strains are highlighted in bold and indicated with circle (●). The scale indicates genetic distance.
Among the Indian strains HPeV1A and HPeV1B, NT identities were found to be in the range 74·09–78·76%. All the 13 HPeV3 strains formed in two separate clusters of which a cluster of five strains showed 91·83–94·36% NT homology with the strains from the Netherlands (GQ183029). Remaining eight strains formed into a separate cluster and showed 77·31–79·31% nucleotide identity with the HPeV3 strains from Japan (AB759191) and the Netherlands (GQ183029). NT homology between these two Indian clusters ranged into 76·3–78·6%. Of the 13 HPeV4 strains, 11 of them clustered closely with Bolivian strains and showed 79·84–81·97% NT identity; other two strains showed highest (93·08–94·43%) NT identity with the strains from France, Sri Lanka and the Netherlands. Two separate clusters of HPeV4 study strains showed 76·46–81·63% NT identity between them. Also, HPeV5 detected in 11 samples, among these 6 strains clustered with the strains isolated in 2004 from the Netherlands (FJ373175) and showed 81·51–83·29% NT identity and remaining cluster of five strains showed 78·88–82·02% NT identity with the Chinese and the Netherlands (EU077511) strains. Two different clusters showed 78·81–82·67% NT identity between them.
The results of deduced amino acid sequence alignment indicated that all the Indian strains contained the highly conserved Q/N and Q or E/S which are believed to be the putative cleavage site of VP3/VP1 and VP1/2A, respectively. All the HPeV1 and 2 strains identified in the present study showed the presence of Arginylglycylaspartic acid (RGD) motif. Interestingly, RGD motif was not found in 7/13 strains of HPeV4 and 2/2 strains of HPeV6 detected.
Clinical features of HPeV genotypes in acute gastroenteritis
HPeV1 was detected predominantly and significantly showed severe infections (30%) as compared with HPeV3, 4 and 5 in acute gastroenteritis cases. HPeV6, 13, 14 and 16 types were found to have severe infections only albeit low frequency of detection. HPeV5 and 11 showed mild to moderate infections. HPeV1 and 3 showed the highest rate of single infection (36% and 11·69%) as compared with HPeV4 and 5 which have been detected predominantly in mixed infections (10·2% and 8·5%). HPeV2 which was detected rarely, HPeV7, 11 and 16 types attributed to mono infections only in comparison with HPeV13 which was found to show mixed infection. Majority of the HPeV1 and 5 infections were detected in children ⩽1 year age group in the acute gastroenteritis cases (Fig. 4).
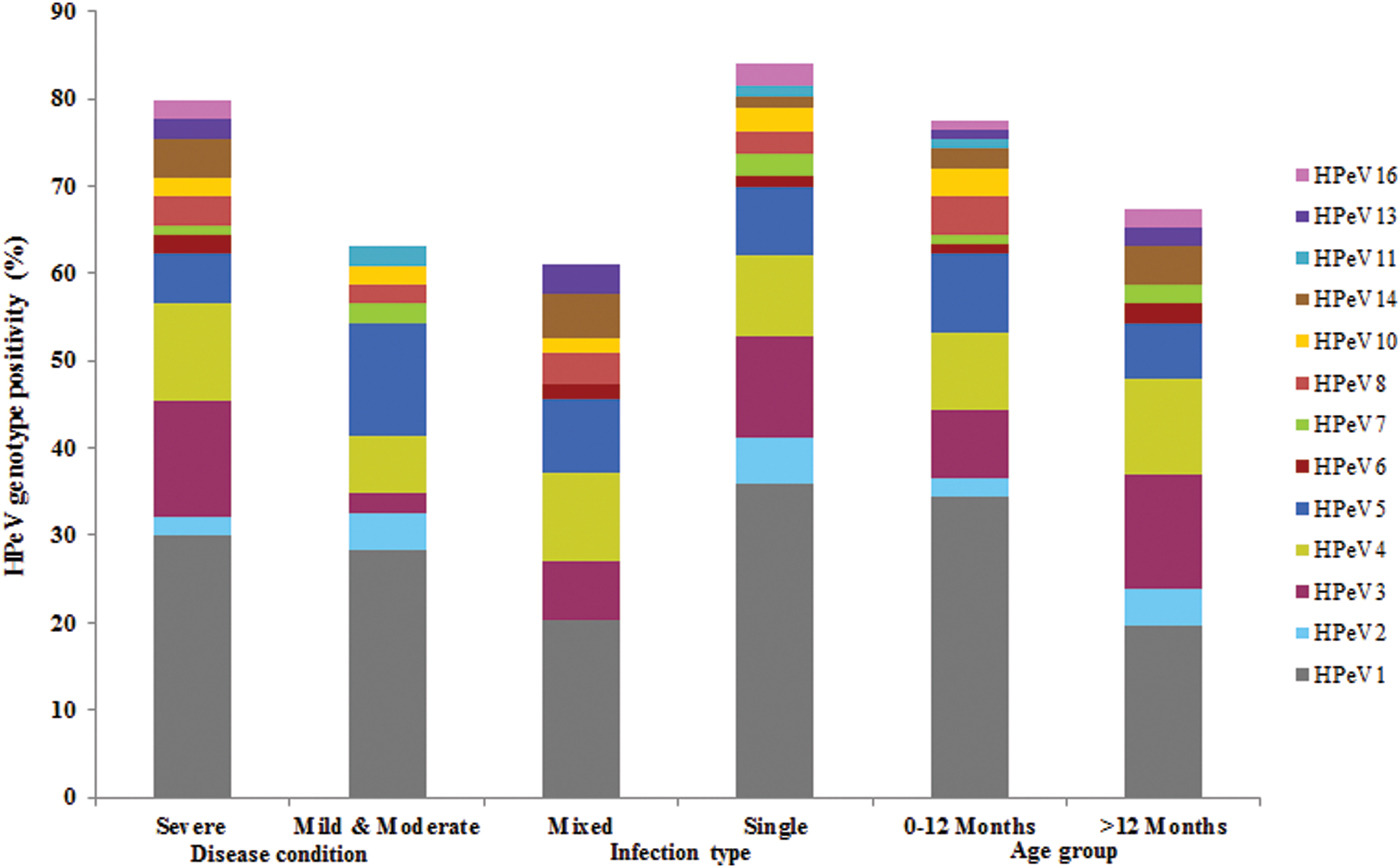
Fig. 4. HPeV Genotype positivity and its correlation with clinical features in acute gastroenteritis patients.
DISCUSSION
HPeVs show a worldwide distribution, however, there have been no strain surveillance studies available on HPeVs in acute gastroenteritis in India. The data generated in this study showed 13·9% of HPeV prevalence in patients with acute gastroenteritis. This is further supported by the studies from other Asian countries reporting 8·1–29·4% HPeV positivity in sporadic cases of acute gastroenteritis [Reference Zhang19, Reference Pham25]. Most of the HPeV infections were detected among the children of <2 years age and the number was highest in the 1st year of life. This is in agreement with the previous studies reported that majority of the HPeV infections were observed below 2 years of age [Reference Baumgarte4, Reference Abed and Boivin26]. Majority of the HPeV infections noted in the study were found in male patients. The studies conducted previously indicated a need for further investigation to understand the difference if any, in susceptibility between the two genders [Reference Ito27].
Co-infections with HPeV and other enteric viruses (43·4%) noted in this study have also been reported from China and Sri Lanka (64·4–66·7%) [Reference Guo, Duan and Qian28, Reference Pham29] suggesting that dual infection is a possible mode of infection. The year wise prevalence of HPeV was reported to be 9·6% and 10·7% in acute gastroenteritis cases during 2006 and 2010 respectively.
In the present study, a total of 13 different HPeV genotypes were detected among Indian children with acute gastroenteritis which suggests circulation of heterogeneous HPeV genotypes. Month-wise distribution of HPeVs in acute gastroenteritis cases could not be analysed as wide range of HPeV types were under circulation and not detected in each month of the year. Therefore, the study has been restricted only to the year wise distribution of HPeVs in acute gastroenteritis cases. Studies reported from other countries have shown that HPeV1 as the predominant genotype in children with acute gastroenteritis [Reference Pham25, Reference Guo, Duan and Qian28, Reference Han30]. Phylogenetic analysis of the HPeV1 sequences demonstrating the existence of two clusters, cluster 1B of the contemporary strains and cluster 1A related to the prototype Harris strain was in agreement with the results of the previous study [Reference Baumgarte4]. The circulation of HPeV1A strains as revealed by our study in India differed from the other Asian countries reporting mainly infections with HPeV1B strains [Reference Zhang19, Reference Pham25, Reference Guo, Duan and Qian28].
In the present study, HPeV3 is the second most predominant genotype detected. The biennial cycle, a distinctive characteristic of HPeV3 has been reported previously [Reference van der Sanden21, Reference Guo, Duan and Qian28]. However, in this study HPeV3 was detected throughout the year. Also, HPeV3 detected previously in younger age group [Reference Benschop31] was found in children of a higher age in the present study which is in agreement with the study reported earlier from China [Reference Zhang19]. Based on the data on HPeV3 occurrence indicates that HPeV3 might have different epidemiological features associated with different geographical regions. In the phylogenetic analysis of full length VP1 gene, eight HPeV3 strains formed in to a separate cluster and also showed 76·3–78·6% NT and 84–86·8% AA identity with other HPeV3 strains which is less than the cutoff value described previously for genotyping of HPeVs [Reference Nix22], could suggest the circulation of HPeV3 variant in Pune, western India.
Interestingly, HPeV2 reported as a rare genotype and mostly in association with gastrointestinal symptoms was identified during 2006–2009 but not in 2010 [Reference Pham32].
Also, failure to genotype 25% of the HPeV strains could be due to mismatching of the primer sets utilised in the study or low titers of virus in the clinical samples.
A Q/N conservation in the VP3/VP1 cleavage site was noted in all the sequences of HPeV strains identified in the study, which corroborated with the report from the previous study and the preference for Glutamine (Q) at the P1 position [Reference Watanabe9]. The C terminal region of VP1 gene contains an RGD motif found essential for replication of HPeV1,2,4,5 and 6 and attachment to cell surface fibronectins αvβ1 and αvβ3 however, other types have been found to lack this surface determinant [Reference Wildenbeest33]. Lack of RGD motif supports the presence of an alternative pathway of cell entry and infection [Reference Alam34]. Also, HPeV4 strains identified in this study showed the circulation of two variants in Indian population one utilising RGD motif and other an alternative RGD independent pathway for cell entry. The occurrence of such variants that belong to the same genotype is common in the family picornaviridae.
Although the role of HPeV in causing acute gastroenteritis has been studied by several investigators, no conclusive data are available to date [Reference Zhang19, Reference Tapia35]. In the present study, HPeVs were detected frequently amongst acute gastroenteritis cases. These data need to be ascertained in acute gastroenteritis patients from wider geographic regions in India for a better understanding of epidemiology and genetic diversity of HPeVs. Further, case-control studies conducted to find out the presence and load of these viruses in the faeces will help to determine their association with diarrhoea.
In summary, the current study identified HPeV genotypes that are circulating in Asian or other countries to date. Also RGD absent’ HPeV5 and 6 strains and co-circulation of two variant strains of HPeV4 in Indian population are reported. The present study highlights the detection and characterisation of a wide spectrum of HPeV genotypes in acute gastroenteritis patients for the first time from India.
ACKNOWLEDGEMENTS
We are grateful to Dr D.T. Mourya, Director, National Institute of Virology, Pune (India) for his constant support. We also acknowledge the cooperation extended by Drs. R. Dhongade, V. Kalrao and A.R. Bavdekar for providing clinical specimens. No external financial support was used in conducting this study.
DECLARATION OF INTEREST
None.








