INTRODUCTION
Shiga toxin-producing Escherichia coli (STEC) are considered to be important foodborne pathogens causing outbreaks and sporadic cases of diarrhoea, haemorrhagic colitis (HC) and haemolytic uraemic syndrome (HUS) in humans worldwide [Reference Bettelheim1]. Cattle and sheep are well established reservoirs of STEC where direct contact with infected animals and consumption of contaminated food and water are important pathways for transmission of STEC to humans [Reference Bettelheim1, Reference Swerdlow2]. Internationally the majority of human cases are associated with O157 STEC [Reference Caprioli3] but non-O157 STEC have also been associated with human disease with six non-O157 serogroups, known as the ‘Super Six’ (O26, O45, O103, O111, O121, O145), responsible for 70% of non-O157 human cases in the USA [Reference Brooks4]. As a consequence of the threat to human health through the contamination of the environment or food products, the Food Safety and Inspection Service (FSIS) of the USA recently extended the list of meat adulterants by including the Super Six STEC [5].
STEC elaborate a variety of virulence factors including Shiga toxin (stx) which inhibits protein synthesis, especially in renal endothelial cells [Reference Louise and Obrig6], E. coli attaching and effacing (eae) and enterohaemolysin (ehxA) genes [Reference Gyles7, Reference Paton8]. The eae gene encodes the outer membrane protein intimin [Reference Jerse9] which mediates the attachment of E. coli to the epithelial lining of intestine and effacement of microvilli [Reference Moon10]. The plasmid-encoded STEC enterohaemolysin, encoded by ehxA, is an important virulence marker which can cause haemolysis of washed sheep erythrocytes [Reference Beutin11]. The exact role of STEC enterohaemolysin in the mechanism of disease is not clearly understood. However, in-vitro studies have shown increased levels of pro-inflammatory cytokine interleukin-1β from human monocytes in response to enterohaemolysin from STEC O128:H12 [Reference Taneike12].
In addition to STEC, cattle and sheep are also reservoirs of enteropathogenic E. coli (EPEC) [Reference Cookson13]. EPEC possess the eae gene and hence form attaching and effacing (AE) lesions on cultured epithelial cells in vitro but lack stx [Reference Kaper14]. The formation of micro-colonies on cultured epithelial cells by many typical EPEC (tEPEC) isolated from human diarrhoeal cases is mediated by the bundle-forming pili (bfpA). However, atypical EPEC (aEPEC) which lack bfpA are commonly recovered from ruminants and may also be associated with human diarrhoeal disease [Reference Trabulsi, Keller and Gomes15].
After the first recorded isolation of STEC in New Zealand in 1980 [Reference Wilson and Bettelheim16] the rate of infection in New Zealand increased to 1·3 cases/100 000 population in 1998 and 3·5 cases/100 000 population in 2011 [17]. In New Zealand the rate of STEC cases in rural areas is three times higher than the rate in urban areas; however, no large outbreaks have been reported to date with most cases being sporadic [Reference Thorbun18]. Shiga toxin bacteriophage insertion typing studies in New Zealand have provided further evidence to support the view that environmental and direct contact pathways are likely to be more important in the transmission of STEC to humans than food pathways [Reference French19]; in line with the strong association observed between cattle density and the incidence of human STEC cases [Reference Thorbun18].
The dairy and meat industries make a significant contribution to the New Zealand economy [Reference Cavanagh and Cavanagh20] and processing methods, especially in the meat industry, are influenced by meat exportation guidelines imposed by some countries such as the USA, which has a zero tolerance policy for STEC O157 and some non-O157 STEC (The Super Six STEC) in meat [5]. Relatively few data are available to assist in the epidemiological analysis of STEC and EPEC in New Zealand cattle. Therefore, this snapshot study was conducted with the aim of isolating E. coli from bobby calves using recto-anal mucosal swabs (RAMS) to determine the distribution of STEC virulence factors (stx1, stx2, eae, ehxA) in randomly chosen isolates. This will also assist in determining whether very young calves may represent a source of STEC or EPEC infection for humans in New Zealand.
MATERIALS AND METHODS
Two abattoirs (A and B) were selected in the North Island of New Zealand and were each visited six times between July and October 2008 [Reference Irshad21]. RAMS (Copan, Italy) were collected systematically from every tenth calf on the chain immediately after slaughter. Twenty-five calves were sampled on each visit and in total 299 calves were sampled.
RAMS were then placed in the transport media provided by the manufacturer and transported to the laboratory on ice where they were enriched in buffered peptone water (BPW) for 24 h at 37°C. Each enrichment broth was serially diluted 10 000 times (10−4) and inoculated onto tryptone bile X-glucuronide (TBX; Fort Richard, New Zealand) and sorbitol MacConkey agar, supplemented with 50 μg/ml cefixime and 2·5 mg/ml potassium tellurite (CT-SMAC) to ensure that single, well-spaced colonies could be subcultured for further analysis. Each plate was then incubated at 37°C for 18–24 h. One blue colony indicative of β-glucuronidase-positive activity and one white colony indicative of β-glucuronidase-negative activity were selected at random from the TBX plates. Similarly, one pink colony indicative of sorbitol-fermenting activity and one grey or colourless colony indicative of sorbitol-non-fermenting activity were selected at random from the CT-SMAC plates. Isolates were subcultured onto the same culture media (CT-SMAC and TBX) to ensure purity, and a single colony was stored at −80°C in nutrient broth containing 15% (v/v) glycerol.
Isolates were analysed using multiplex polymerase chain reaction (PCR) to detect stx1, stx2, eae and ehxA genes. Isolates were re-grown on TBX or CT-SMAC at 37°C for 18–24 h. A single colony was suspended in 500 μl of 2% chelex (Bio-Rad, New Zealand) solution and heated at 95°C for 10 min for isolation of DNA. The lysed bacterial cell suspension was then cooled in the refrigerator for 2 min and re-centrifuged at 12 000 g for 2 min. The supernatant containing the DNA was transferred to another tube. Each PCR reaction contained 1x reaction buffer (Invitrogen, New Zealand), 0·2 μ m of each primer [Reference Paton and Paton22], 0·1 mm of each dNTP (Fermentas, New Zealand), 0·15 mm MgCl2 (Invitrogen), 1 U Taq DNA polymerase (Invitrogen), 2 μl DNA, and made to a final volume of 25 μl with sterile water. The amplification was carried out in a Gene Amp PCR system 9700 (Applied Biosystems, Australia). The cycling conditions for multiplex PCR were: an initial denaturation step for 5 min at 96°C followed by 40 cycles of 30 s at 96°C, 30 s at 60°C, 30 s at 72°C, with a final extension of 5 min at 72°C, after which the PCR products were electrophoresed through an agarose (2% w/v) gel (Invitrogen) and visualized using ethidium bromide under ultraviolet illumination.
Isolates positive for the targeted virulence genes were confirmed as being E. coli by PCR targeting the alanine racemase gene [Reference Yokoigawa23]. Briefly, each PCR reaction contained 1x reaction buffer (Invitrogen), 0·2 μ m of each primer (forward: 5′-CTGGAAGAGGCTAGCCTGGACGAG-3′; reverse: 5′-AAAATCGGCACCGGTGGAGCGATC-3′), 0·1 mm of each dNTP (Fermentas), 0·15 mm MgCl2 (Invitrogen), 1 U Taq DNA polymerase (Invitrogen), 2 μl of DNA, and made to a final volume of 20 μl with sterile water. The amplification was carried out in a Rotor Gene 6000 series thermal cycler. It was programmed for an initial denaturation step of 2 min at 94°C, 25 cycles of 20 s at 94°C, 20 s at 62°C, and 20 s at 72°C and a final extension at 72°C for 90 s. The PCR products were electrophoresed through an agarose gel (1% w/v) and visualized by staining the gel with ethidium bromide under ultraviolet illumination.
A PCR for the detection of bfpA encoding the EPEC major bundle-forming pilus subunit was performed on all eae-positive, stx-negative isolates. PCR primers bfpF (5′-GAAGTAATGAGCGCAACGTCTGC-3′) and bfpR (5′-GGTAAGYGTCAGATAGTAACC-3′) were designed using the bfpA sequence from EPEC strain O127:H6 E2348/69 (Genbank accession no. Z68186) to amplify a 213 bp DNA product. A degenerate base Y was included in the reverse oligonucleotide primer to account for a single nucleotide polymorphism observed at that position in other bfpA sequences during primer specificity analysis. Each PCR reaction contained 0·1 μ m of each primer, 17·5 μl PCR Platinum Supermix (Invitrogen) and 1·5 μl template DNA. EPEC strain O127:H6 E2348/69 was used as the positive control strain. The amplification was carried out in a Master Cycler proS (Eppendorf AG, Germany) thermal cycler with an initial denaturation step of 2 min at 96°C, followed by 30 cycles of 96°C for 30 s, 53°C for 30 s, and 72°C for 60 s and a final extension of 72°C for 10 min. PCR products were viewed as described previously.
Isolates positive for any of the targeted virulence factors were analysed by real-time PCR (qPCR) to amplify genes that are diagnostic for E. coli serogroups O26 [Reference Perelle24], O103 [Reference Fratamico25], O111 [Reference Perelle24], O145 [Reference Fratamico26] and O157 [Reference Perelle24]. STEC isolates negative for E. coli O26, O103, O111, O145 and O157 by qPCR were serotyped by the Enteric Reference Laboratory, ESR, New Zealand using O and H antigens (Statens Serum Institute, Denmark). Sixty isolates containing the targeted virulence genes were genotyped using pulsed-field gel electrophoresis (PFGE) to determine the genetic relatedness of isolates. PFGE was performed following the PulseNet USA protocol [27]. The gel was stained in ethidium bromide (0·5 μg/ml) for 20 min and visualized using short wavelength ultraviolet transillumination. Digital images of the gel were captured using a Gel Doc imaging system (Bio-Rad, Italy) and saved as TIFF files. The digital images were analysed using Bionumeric v. 6.6 (www.applied-maths.com). The unweighted pair-group method with arithmetic mean (UPGMA), Dice coefficient with >70% similarity cut-off, 0·5% optimization and band-matching tolerance were used to construct the clusters of E. coli isolates with one or more targeted virulence genes.
The distribution of farms from which the calves that were sampled originated from (n = 193) were mapped using R package maptools (R Development Core Team, R Foundation for Statistical Computing, Austria). The coordinates (latitude and longitude) of the farms (n = 180) were obtained using Agribase data [Reference Sanson and Pearson28] and Google earth (www.google.com/earth). The latitude and longitude of each farm was then converted to x,y coordinates. The coordinates of 13 farms were missing. The sampled farms were aggregated to 5 × 5 km grid squares to maintain the anonymity of the farms. The prevalence of calves positive for E. coli isolates with one or more targeted virulence genes in three regions (Manawatu, Taranaki, Waikato) of the North Island of New Zealand was estimated and adjusted for the lack of independence of calves within farms using R package ‘survey’ and function ‘svydesign’.
RESULTS
In total 299 RAMS were collected from bobby calves originating from 193 farms in the North Island of New Zealand. Microbial isolates (n = 975) obtained from RAMS were analysed by multiplex PCR and of these, 512 isolates were obtained from CT-SMAC (299 isolates were sorbitol fermenters and 213 were sorbitol non-fermenters) and 463 from TBX agar (227 isolates were β-glucuronidase positive and 236 isolates were β-glucuronidase negative). Sixty of the 975 isolates were positive for at least one of the targeted virulence factors of which 47/512 (9·1%) were isolated on CT-SMAC. Of these, a higher proportion of sorbitol fermenters (42/299, 14%) were positive for the targeted virulence factors compared to sorbitol non-fermenters (5/213, 2·3%) (χ 2 test, P = 0·00001). Thirteen (2·8%) of the 463 isolated on TBX were positive for the targeted virulence factors. Of these 9/227 (3·9%) of the β-glucuronidase-positive isolates were positive for the targeted virulence factors compared to 4/236 (1·7%) of the β-glucuronidase-negative isolates (χ 2 test, P = 0·23). The rate of isolation of E. coli isolates with one or more targeted virulence genes was significantly higher on CT-SMAC (47/512, 9·1%) compared to TBX (13/463, 2·8%) (χ 2 test, P < 0·0001). The most common combination of the targeted virulence genes in these isolates was eae, ehxA (n = 35) followed by eae (n = 9); ehxA (n = 8); stx2, eae, ehxA (n = 4); stx1, eae, ehxA (n = 3) and stx2 (n = 1) (Table 1). None of the eae-positive stx-negative strains (n = 44) were bfpA-positive. Two of the three stx1, eae, ehxA isolates were identified as serogroup O26 using qPCR and the third strain as O71:HR using traditional serological methods. Three of the four stx2, eae, ehxA isolates were identified as O157 using qPCR and the fourth as serogroup O68:H24. The stx2-positive isolate was identified as ONT:HNM. Six of the eight stx-positive isolates were detected on CT-SMAC and two on TBX (χ 2 test, P = 0·35). In total, 53/299 (17·7%) calves were positive for isolates possessing one or more of the targeted virulence genes (stx1, stx2, eae, ehxA). Of these 2·6% (8/299) calves were positive for STEC, 12·3% (37/299) for aEPEC and 2·6% (8/299) for ehxA. Isolates positive for eae, stx or ehxA or combinations thereof were obtained from the same calf RAMS enrichment broth on seven different occasions (Fig. 1). Isolates having contrasting serogroups were obtained from four of the seven aforementioned RAMS enrichments; serogroup O103 and O145 (n = 2), serogroup O26 and O145 (n = 1) and serogroup O145 and an untyped isolate (n = 1). On all four of these occasions each isolate was eae- and ehxA-positive only. Untyped isolates EcCa419a and EcCa419b were eae- and ehxA-positive only but had contrasting PFGE profiles. In contrast to serogroup O145 isolate EcCa388a, that was eae- and ehxA-positive, an untyped isolate (EcCa388c) from the same animal was ehxA-positive only. Untyped strains EcCa412a and EcCa412c were clustered together but appeared to have contrasting PFGE profiles. Finally, serogroup O145 isolates EcCa237a and EcCa237c possessed very similar PFGE profiles and were clustered together indicating that they may have been of a similar genotype.

Fig. 1. Clustering using UPGMA and Dice coefficient of the PFGE profiles of E. coli isolates positive for stx1, stx2, eae and ehxA from calves with >70% similarity cut-off using XbaI. The last lane is the Salmonella serotype Braenderup reference standard (H9812). Isolates with the same EcCa numbers were obtained from the same calves.
Table 1. Distribution of the targeted virulence genes (stx1, stx2, eae, ehxA) in E. coli isolates obtained from recto-anal mucosal swabs (n = 299) of bobby calves on sorbitol MacConkey agar (CT-SMAC) and tryptone bile X-glucuronide (TBX) plates. Sorbitol-fermenting (SF) and non-sorbitol-fermenting (NSF) colonies were obtained on CT-SMAC plates while β-glucuronidase-positive (βGP) and β-glucuronidase-negative (βGN) colonies were obtained on TBX
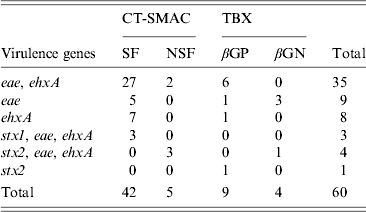
There was no significant difference in the proportion of calves positive for E. coli isolates with one or more targeted virulence genes from Waikato [23·1%, 95% confidence interval (CI) 11·9–40], Manawatu (21·1%, 95% CI 13·1–32) and Taranaki (15·7%, 95% CI 10·3–23) regions (Table 2). Using qPCR, three of the 35 eae-, ehxA-positive isolates were identified as serogroup O26, seven as O103 and eight as O145. Similarly, using qPCR three of the nine eae-positive isolates were identified as serogroup O26. None of the remaining 23 isolates gave a positive result with O26-, O103-, O111-, O145- and O157-specific qPCR primers and their respective serogroups were not determined.
Table 2. The proportion of recto-anal mucosal swabs positive for E. coli isolates containing the targeted virulence genes (stx1, stx2, eae, ehxA) collected from 299 bobby calves from three regions of the North Island of New Zealand
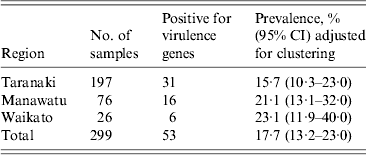
CI, Confidence interval.
The distribution of the farms from which calves harbouring E. coli were obtained in this study is shown in the Figure 2. Isolates positive for the targeted virulence genes were found in all important dairy producing areas of the North Island of New Zealand from which calves were sampled.
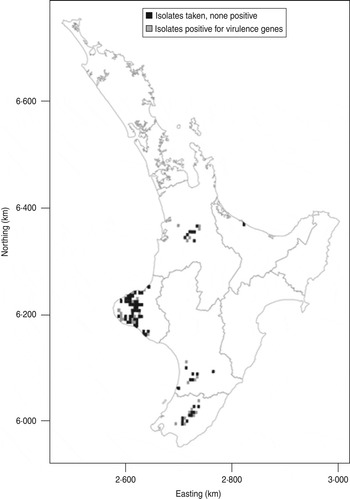
Fig. 2. Map showing distribution of E. coli isolates positive for the targeted virulence genes (stx1, stx2, eae, ehxA) in the North Island of New Zealand. The data are aggregated to 5 × 5 km grid cells. If an animal was positive for any of the targeted virulence genes, the grid cell is grey. If the animals sampled were negative the cell is black.
Using XbaI–PFGE, the 60 isolates from which at least one STEC-associated virulence factor was detected, could be divided into 15 clusters (A–O) with >70% similarity cut-off (Fig. 1). Five of the eight O26 isolates grouped in cluster L; another O26 isolate (eae-positive only) was in cluster B and two eae-positive O26 isolates could not be genotyped using XbaI–PFGE. All seven O103 isolates grouped in cluster I. Of eight O145 isolates, seven were grouped in cluster I and one isolate belonged to cluster O. All O157 isolates (n = 3) grouped together in cluster M. All O103 isolates present in cluster I were obtained from Taranaki (n = 6) and Manawatu (n = 1) regions. Of six O103 isolates from Taranaki four were recovered from one farm and the remaining two from two different farms. All O145 isolates in cluster I were obtained from Taranaki from four different farms. Similarly, four of the O26 isolates in cluster L were obtained from four different farms in Taranaki and the one isolate from Manawatu region. All O157 isolates in cluster M were recovered from three different farms in Taranaki region.
DISCUSSION
Cattle have been identified as a reservoir for both O157 STEC and non-O157 STEC and may provide a source of human infection through direct animal contact and faecally contaminated material, or through environmental contamination. This study has provided evidence that young calves are potentially important carriers of STEC and aEPEC in the North Island of New Zealand. The results of this study should be extrapolated with care as samples were collected from only two abattoirs and the prevalence of STEC and/or aEPEC (bfpA-neagtive EPEC) in other areas of the North Island of New Zealand may be influenced by variables such as the concentration of dairy farms and climatic conditions. As this study aimed to provide a snapshot of the distribution of targeted virulence factors, it is highly likely that the prevalence of both STEC and aEPEC described in this study are underestimates as no preliminary screening method such as an initial screen for stx was used. However, the use of two culturing media, selection of four randomly chosen colonies with an absence of screening of enriched samples for stx have provided an opportunity to retrieve eae- and/or ehxA-positive isolates.
A low proportion of STEC (2·3%) isolates were recovered in this snapshot study with seven stx- and eae-positive strains and a single stx-only isolate obtained. These data are in contrast to a previously described study conducted in the North Island of New Zealand where STEC (n = 139) were isolated from 53/187 (28·3%) RAMS samples taken from cattle (calves, heifers, dairy cattle) [Reference Cookson, Taylor and Attwood29]. Furthermore, the prevalence of stx-only isolates (48/139, 34·5%) was higher than the prevalence of both stx- and eae-positive (21/139, 15·1%) isolates and the remaining 70 isolates (50·3%) were eae-positive only [Reference Cookson, Taylor and Attwood29]. Importantly, RAMS were taken from animals of different age groups (calves, heifers, dairy cattle) from four farms in two regions of the lower North Island in the Cookson et al. [Reference Cookson, Taylor and Attwood29] study. A study of STEC in Spanish cattle where faecal samples were collected from calves, heifers and adult cattle over a period of 1 year reported a higher prevalence (8·7%, 36/412). These samples were inoculated onto MacConkey agar and four E. coli colonies from each plate were analysed for the presence of stx, eae and ehxA genes [Reference Orden30].
STEC strains of zoonotic importance [Reference Bettelheim1] belonging to serogroups O26, O68, O71 and O157 isolated in this study have previously been isolated from healthy cattle and sheep in New Zealand, Australia and Spain [Reference Bettelheim1, Reference Cookson13, Reference Orden30]. Further, STEC strains from these serogroups, have been isolated from human cases [Reference Bettelheim1] with STEC O157 and O26 frequently isolated from ruminants and human cases but STEC O68 and O71 only rarely isolated from these hosts [Reference Bettelheim1]. Bettelheim [Reference Bettelheim1] reported isolation of O68 from healthy cattle and human cases on three and seven occasions, respectively. Similarly, O71 have also been isolated from healthy sheep and human cases on two and three occasions, respectively. STEC O71 (stx1- and stx2-positive) has also been isolated from a diarrhoeic lamb [Reference Orden31].
The zoonotic potential of these serogroups should not be underestimated for two reasons. First, the low isolation of these serogroups may be due to unavailability of standardized isolation methods and second, the recent emergence of STEC O104:H4 in Europe affecting 3222 individuals including 810 HUS cases has shown that less virulent STEC can become highly virulent by acquiring other virulence factors [Reference Frank32].
In our study all STEC O157 isolates were positive for the stx2 gene whereas all STEC O26 isolates were positive for the stx1 gene. Previous studies have also reported higher prevalence of stx2 in STEC O157 isolates [Reference Irshad21] and stx1 in STEC O26 isolates [Reference Cookson13]. For example, all STEC O157 isolates (n = 10) obtained from New Zealand calves were stx2-positive [Reference Irshad21]. Similarly, Cookson et al. [Reference Cookson13] reported that all O26 isolates (n = 2) obtained from cattle were positive for the stx1 gene.
Previous studies have shown an association between stx genotype and STEC virulence. STEC isolates with the stx2 gene are considered to have high virulence due to their association with HC and HUS cases [Reference Boerlin33]. Boerlin et al. [Reference Boerlin33] reported that the rate of isolation of stx2-positive isolates was higher (39/60, 60%) from HUS and HC cases than stx1-positive isolates (29/75, 38·6%). Another study also found that 12/15 (80%) STEC isolates obtained from HUS and HC cases were positive for stx2, one for stx1 and stx2 (6·6%) and the remaining two for stx1 (13·3%) [Reference Pradel34].
STEC O26 is also an important human foodborne pathogen and has caused large outbreaks of HUS where the vehicle of infection was food [Reference Hiruta, Murase and Okamura35]. In contrast to much of the rest of the world, in central and southern Europe the numbers of human cases of diarrhoeal disease due to STEC O26 are much higher [Reference Tozzi36]. However, in New Zealand very few human cases due to STEC O26 have been reported and this may be due to underreporting of STEC cases where O26 is not isolated or identified on selective media [Reference Baker37]. Most of the STEC O26 that are associated with human diarrhoeal disease possess stx1, eae and ehxA. In this New Zealand study a typical virulence profile of stx1, eae and ehxA was noted for all the STEC O26 isolates. This virulence profile has public health significance as this is the most common virulence profile observed in STEC O26 isolates from human cases internationally [Reference Zhang38].
The proportion of calves positive for aEPEC (12·7%) in this study was estimated to be higher than that of STEC (2·3%). However, there was no difference in prevalence of STEC and aEPEC in cattle in other studies [Reference Cookson13, Reference Orden30]. As for variation with STEC prevalence noted in this study compared to previous work, any variations may be due to differences in sampling and analysis of samples.
Most of the EPEC isolated from ruminants in previous studies lack bfpA and are therefore considered as aEPEC [Reference Trabulsi, Keller and Gomes15]. The provenance of aEPEC is uncertain as several studies have shown that aEPEC may eventuate from STEC strains that have lost the stx-encoding bacteriophage, or where the complete E. coli adherence factor (EAF) plasmid, or the operon encoding bfpA on the EAF plasmid has been lost [Reference Bielaszewska39, Reference Hernandes40]. In contrast, the acquisition of the stx-encoding bacteriophage by aEPEC has been reported, suggesting that transition to and from aEPEC and STEC is a dynamic process in complex microbial environments such as ruminant or human gut [Reference Whittam41]. To our knowledge there are no instances of isolates being bfpA- and stx-positive. As isolates positive for bfpA and stx have not been recognized, the classification of eae-positive isolates remains unclear as some may be tEPEC (bfpA-positive), others may be aEPEC (EAF plasmid lost) or STEC-like (stx genes lost). The prevalence of aEPEC (37/299, 12·3%) in this study was lower than that of a previous study (44% in calves) conducted in the lower North Island of New Zealand [Reference Cookson13] but may be due to contrasting study designs. Some of the aEPEC serogroups (O26, O103, O145) isolated in this study may be of zoonotic importance due to their strong association with HC and HUS cases [Reference Bielaszewska39].
Although ehxA is considered an important virulence marker of STEC due to its association with disease in humans [Reference Schmidt, Beutin and Karch42], the role of plasmid-associated enterohaemolysin in human disease is not clearly understood. Most human clinical cases of disease caused by STEC are associated with strains that are eae- and ehxA-positive [Reference Gyles43]; however, some serotypes such as O113:H21 (eae-negative, ehxA-positive), are notable exceptions [Reference dos Santos44]. In this study six of the seven isolates were positive for eae and ehxA indicating their potential for causing disease in humans. However, STEC isolates without eae and ehxA have been recovered from diarrhoeal and HUS patients on rare occasions [Reference Beutin and Martin45]. The virulence of eae- and ehxA-negative isolates should not be underestimated. The recent European outbreak of STEC O104, involving isolates negative for eae and ehxA further illustrates the potential public health significance of STEC isolates negative for eae and ehxA. Most of the aEPEC (35/44, 79·5%) in the present study were also positive for ehxA. Another study also reported higher detection of plasmid-associated genes such as ehxA, etpD, espP and katP in aEPEC (40/80, 50%) compared to tEPEC (6·9%) [Reference Bugarel46].
CT-SMAC has been frequently used as a selective media for the isolation of STEC O157. However, a previous study has also reported the isolation of STEC serogroups O5, O84, O128 and various non-typable STECs on CT-SMAC [Reference Cookson13]. Furthermore, in this study, the STEC O68:H24 strain was isolated on CT-SMAC. Results from this study indicated a higher rate of isolation of E. coli isolates with the targeted virulence genes on CT-SMAC compared to TBX (χ 2 test, P < 0·0001) which highlights the benefit of using two different media. Similarly, sorbitol-fermenting E. coli isolates were more commonly positive for the targeted virulence genes compared to non-sorbitol-fermenting E. coli isolates (χ 2 test, P = 0·00001) indicating the importance of analysing two different colonies on CT-SMAC. A previous study has suggested that some E. coli strains, especially those STEC that are stx-positive and eae-negative, are unable to grow on CT-SMAC [Reference Fukushima, Hoshina and Gomyoda47], therefore an additional medium, TBX, was included to enhance the breadth of E. coli that were isolated. The random selection of four colonies (blue, white, grey, purple) provides at least two (potentially four) separate isolates to study in further detail for the distribution of targeted STEC virulence factors. Confining preliminary selection to a single agar medium and/or fermentation characteristic may have limited the breadth of observed allelic profiles recorded. In the study by Cookson et al. [Reference Cookson13] most eae-negative STEC were isolated from TBX (38/48, 79·16%) compared to CT-SMAC (10/48, 20·84%). However, in this study, where very young calves were sampled, only one eae-negative STEC (ONT:HNM) was isolated from TBX. Similarly, in this study aEPEC were isolated more frequently on CT-SMAC (n = 35) compared to TBX (n = 10). Cookson et al. [Reference Cookson, Taylor and Attwood29] also reported higher isolation of aEPEC on CT-SMAC (67/70: 41 sorbitol-fermenting, 26 non-sorbitol-fermenting) compared to TBX (3/70). Although it is difficult to compare those studies due to differences in study design, these findings suggest the possibility of underestimation of STEC or EPEC prevalence if a single selective medium is used for isolation of E. coli.
PFGE is a molecular epidemiology tool and has been used for subtyping STEC [Reference D'Costa48]. In this study isolates having the same serogroup generally clustered together with few exceptions. For example, one E. coli O145 isolate grouped separately from the remaining seven O145 isolates and one O26 from the remaining O26 isolates. The basis of this differential clustering is unknown. It may be due to the presence of some additional virulence genes which are absent from other isolates. The presence in the same PFGE cluster of similar isolates obtained from different farms in the same region provides evidence of localized geographical clustering of farms positive for various E. coli serotypes. For example, O103 isolates obtained from different farms in the same region grouped together by PFGE. A similar trend was observed for O26, O145 and O157 isolates. This geographical clustering may be due to localized transmission of various E. coli serogroups between farms in the same region.
This study indicates that STEC and aEPEC of public health significance are present in the bobby calves in New Zealand and therefore, they may represent an important source of environmental contamination and possible human infection. However, studies using molecular characterization and comparison of these isolates with isolates from human cases are required to estimate the nature and potential risk to the human population from these E. coli. Additional non-O157 serogroups (O26, O45, O103, O111, O145) have recently been legislated as adulterants by FSIS [5]. Some of these serogroups were isolated in this study where both stx-positive and stx-negative strains were characterized. Other non-O157 serogroups may have been missed with this snapshot study and more targeted methods such as qPCR or immunomagnetic separation will be required to unequivocally determine their presence.
ACKNOWLEDGEMENTS
We thank the New Zealand Foundation for Research, Science and Technology for financially supporting this work which contributes to the IMPACT project (contract no. C03X0701). We also thank the Higher Education Commission of Pakistan for providing funds for this study.
DECLARATION OF INTEREST
None.







