Introduction
Chikungunya (CHIKV) and Dengue (DENV) viral infections are prevalent in many geographical regions, especially in the tropical and sub-tropical regions. CHIKV has caused approximately 2.5 million infections over the past decade and was identified in around 40 countries in the Indian subcontinent, Africa, Latin America, South-East Asia, USA and Europe [Reference Staples and Fischer1–Reference Sharp3]. DENV circulates in more than 125 countries and around 390 million cases are reported annually [Reference Bhatt4]. One of the major factors for co-circulation and co-infection with CHIKV and DENV in many geographical regions is due to the common vector (Aedes aegypti and Aedes albopictus mosquito) [Reference Ferreira-de-Lima and Lima-Camara5, Reference Chang6]. Both the viral infections show similar clinical presentations such as high fever, headache, nausea, vomiting, rashes, arthralgia and myalgia. Some unique clinical manifestations include low platelet count, haemorrhage and shock syndrome in DENV whereas sudden and severe muscle spasm including intense joint pain in CHIKV fever [Reference Nazish7]. CHIKV is divided into three genotypes: ECSA (East Central South African), Asian and West African. DENV has four serotypes (DENV 1–4) and these are subsequently divided into four to five genotypes.
New Delhi situated in northern part of India is hyper-endemic for DENV infection since all the four serotypes have been identified [Reference Islam8–Reference Bharaj11]. Heavy rainfall, high humidity, rapid urbanisation, high population density, inadequate drainage system and fluctuating climate scenarios contribute towards mosquito proliferation in this region. This leads to regular outbreaks of mosquito-borne infections (CHIKV, DENV and malaria fever). Delhi has previously reported many CHIKV cases [Reference Afreen10, Reference Kaur12–Reference Shrinet14] and DENV outbreaks (1967, 1970, 1982, 1988, 1996, 2003, 2006 and 2010) [Reference Balaya15–Reference Kumari, Kumar and Chauhan22]. In 2016, another major outbreak of DENV along with CHIKV viral infection took place in New Delhi with a predominance of CHIKV cases [Reference Nazish7, Reference Kaur12]. The National Vector Borne Disease Control Programme (NVDCP) reported 12,279 cases of CHIKV and 4431 cases of DENV infection from New Delhi during 2016 [23, 24].
The present study was planned to identify and characterise the CHIKV and DENV in the samples collected from symptomatic patients from the local health centre in New Delhi during 2016. The viruses were detected in the clinical samples by RT-PCR and the strains were characterised by phylogenetic, mutational, selection pressure and entropy analysis. Characterisation of the co-circulating strains of CHIKV and DENV is envisaged to delineate the evolutionary trajectory of these arboviruses that is likely to contribute towards the formulation of effective control measures.
Methods
Collection of clinical samples
The patients attending the Out-Patient Department (OPD) of Dr. M.A. Ansari Health Centre, Jamia Millia Islamia, New Delhi were enrolled for the study. Acute phase blood samples from the patients with symptoms of CHIKV-like or DENV-like illness were collected during August to November 2016. The clinical and demographic information of the patients were collected in proformas. The study was approved by the Institutional Ethics Committee, Jamia Millia Islamia, New Delhi. Written informed consent was obtained from the enrolled patients before the sample collection. Around 2–3 ml of blood was collected from the patients by a trained medical professional. The blood samples were transported to the virology laboratory. The sera were separated by centrifugation of the samples at 10 000 rpm for 5 min. The serum was aliquoted and stored at −80 °C.
IgM-Screening
The 65 blood samples collected from the patients with CHIKV-like illness were tested for CHIKV IgM by rapid card test (OnSite CHIKV IgM combo Rapid Test kit, CTK Biotech, USA) as per the manufacturer's instructions.
RNA extraction and cDNA synthesis
RNA was extracted from all the serum samples using RNA Sure Virus kit (Genetix, India) as per the manufacturer's instructions. The cDNA was synthesised from RNA using High Capacity cDNA Reverse Transcription Kit (Applied Biosystems, USA) according to the manufacturer's instructions. The cDNA was used as a template for amplification of both CHIKV and DENV specific genes in the clinical samples.
Detection of CHIKV and DENV in clinical samples
CHIKV and DENV were identified in the samples using published primer sequences [Reference Lanciotti25, Reference Santhosh26]. PCR for the E1 gene was used for identification of CHIKV in the clinical specimens. Similarly, the external and semi-nested PCR reaction for the CprM region was used for detection of DENV in the clinical samples. The cycling conditions of both the PCR reactions were standardised in our laboratory [Reference Afreen10]. The size of the CHIKV and DENV-3 amplicons was 852 and 290 bp, respectively. The amplicons were resolved on two percent agarose gel and visualised in UV-light using gel documentation system (Wealtec Sparks, NV, USA).
DNA sequencing and its analysis
DNA sequences of the partial E1 gene of CHIKV and CprM region of DENV were determined in the present investigation. The PCR products were excised from the agarose gel and DNA was extracted using Gel Extraction Kit (Plus DNA Clean/Extraction Kit, GeneMark, Taiwan) as per the manufacturer's instructions. The amplicons were sequenced in forward and reverse directions using commercial services (Applied Biosystem, USA). The nucleotide ambiguities were resolved and sequences were manually edited in GeneDoc (v2.7) and BioEdit (v.7.2). All the CHIKV and DENV study sequences were confirmed by online BLAST tool at NCBI. The mutations in the CHIKV and DENV-3 study strains were analysed with respect to the prototype strain.
Phylogenetic analysis
The GenBank sequences were downloaded and aligned with the study sequences in BioEdit. The phylogenetic trees were generated by the Neighbour-Joining and Maximum-Likelihood method in MEGA6 (v.6.06). The tree robustness was assessed with 1000 replicas. The genetic distances among the sequences were calculated with Tamura Nei method of nucleotide substitution. The following prototype strains were used for the analysis: S27 strain of CHIKV (Accession number AF 369024) and H87 strain of DENV-3 (Accession number M93130).
Selection pressure analysis
Selection pressure analysis in the E1 gene of CHIKV and CprM region of DENV was investigated at individual codons position using Datamonkey web-server (http://www.datamonkey.org). The ratio of non-synonymous to synonymous mutations (dN/dS) was calculated using five different analysis methods, single likelihood ancestor counting (SLAC), internal fixed effects likelihood (IFEL), random effects likelihood (REL), mixed effects model of evolution (MEME) and fixed effects likelihood (FEL) by utilising HKY85, F81 and REV method of nucleotide substitution. If the ratio of dN/dS is less than 1, it is known as negative or purifying selection. If the value is equal to 1, it is known as neutral selection. If the ratio is more than 1 it is known as positive selection. The sites identified to be under positive selection by at least two different methods were considered as the positively selected sites with P-value between 0.05 and 0.25.
Shannon entropy analysis
BioEdit (v.7.2.) software was used for the Shannon entropy analysis. It is used for the identification of variable sites at particular amino acid positions. High entropy value indicates a higher possibility of variation. The data collected from the online software through various entropy calculations were then transferred into excel sheet to plot the Shannon entropy graph.
Results
Characteristics of the patients
Patients with suspected CHIKV-like or DENV-like illness were recruited from the OPD of the Health Centre, Jamia Millia Islamia from August to November 2016. A total of 130 blood samples were collected from the patients. The clinical and demographic details of the patients are listed in Supplementary Table S1. High-grade fever (103–105 °C) was observed in all the patients. Other clinical manifestations of the patients with CHIKV-like illness include a headache, nausea, facial and pedal swelling, hyperpigmentation, sudden muscle spasm and intense joint pain with inability to move in many patients. The patients with DENV-like illness presented with additional symptoms of body ache, muscle pain, headache, retro-orbital pain, nausea, vomiting and rashes.
Detection of CHIKV and DENV infection by RT-PCR
All the samples collected during the study were subjected to RT-PCR for CHIKV and/or DENV. Sixty-five samples were tested for CHIKV and 120 samples for DENV, depending on the clinical symptoms of the patients. Thus, a total of 55 samples were tested for both the viruses. CHIKV was identified in 26 samples (40%) of the 65 samples tested. Similarly, DENV was detected in 48 samples (40%) of the 120 samples tested. Concurrent infection with both the viruses was detected in five (9%) samples (Fig. 1). Two samples showed concurrent infection by three arthropod mediated pathogens, namely, DENV-3, CHIKV and Plasmodium vivax [Reference Tazeen27, Reference Abdullah28].
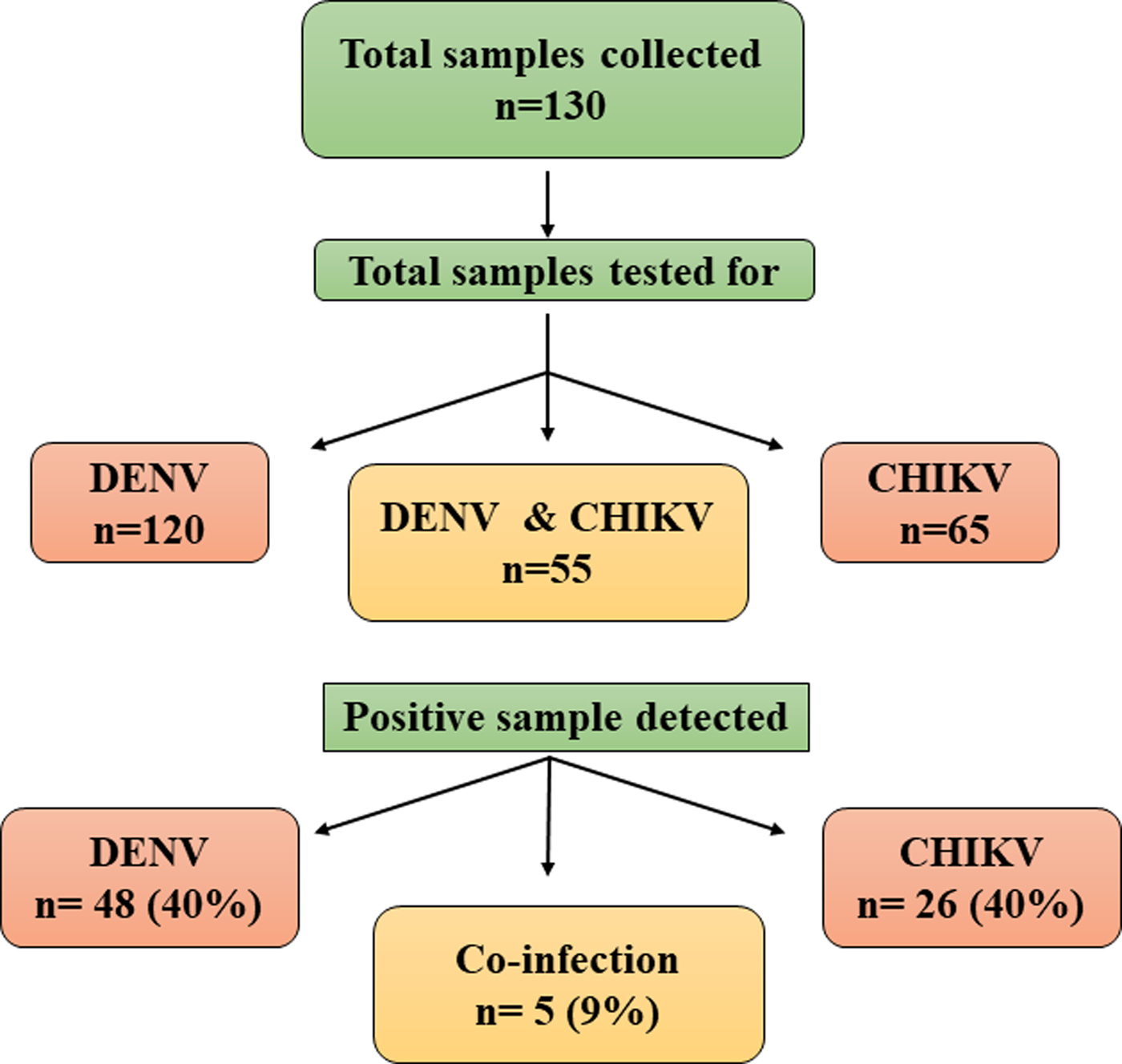
Fig. 1. Schematic representation of samples analysed for Chikungunya and Dengue viruses.
IgM-screening for CHIKV
All the 65 samples collected from the suspected CHIKV patients were tested for IgM antibodies with rapid card test (OnSite CHKV IgM combo Rapid Test kit). Twenty-one samples were positive for CHIKV IgM of the samples tested.
Correlation of demographic and clinical features with the viral infection
A correlation between CHIKV and DENV with their demographics is described in Tables 1 and 2. Fever was the most common clinical manifestation observed in all the suspected patients. The age of symptomatic patients was between 3 and 75 years. The patients were divided into four different age groups i.e. 0–20, 21–40, 41–60 and >61 years. The viral infection (CHIKV or DENV) was correlated with the demographic and clinical features.
Table 1. Demographic and clinical information of CHIKV strains

Table 2. Demographic and clinical information of DENV strains

Of the suspected CHIKV patients (n = 65), 46 (70.76%) were males and 19 (29.24%) were females with a male to female ratio of 1:0.41. Mean age of suspected patients was 31.1 years (s.d. ± 14.1) and mean duration of illness was 3.8 days (s.d. ± 1.4). The mean age for CHIKV positive patients [Reference Santhosh26] was 34.7 years (s.d. ± 17.7) and mean duration of illness was 4.15 days (s.d. ± 0.19) with a male to female ratio of 1:0.3. The age-wise analysis of the data suggested that most of the positive patients (15) belonged to 21–40 years age group followed by three patients in 0–20, four patients in 41–60 and four patients in >61 age group. Sixty-nine percent of the symptomatic patients had joint pain, 52% had muscle pain and 47% had swelling (Fig. 2a, Tables 1 and 3).
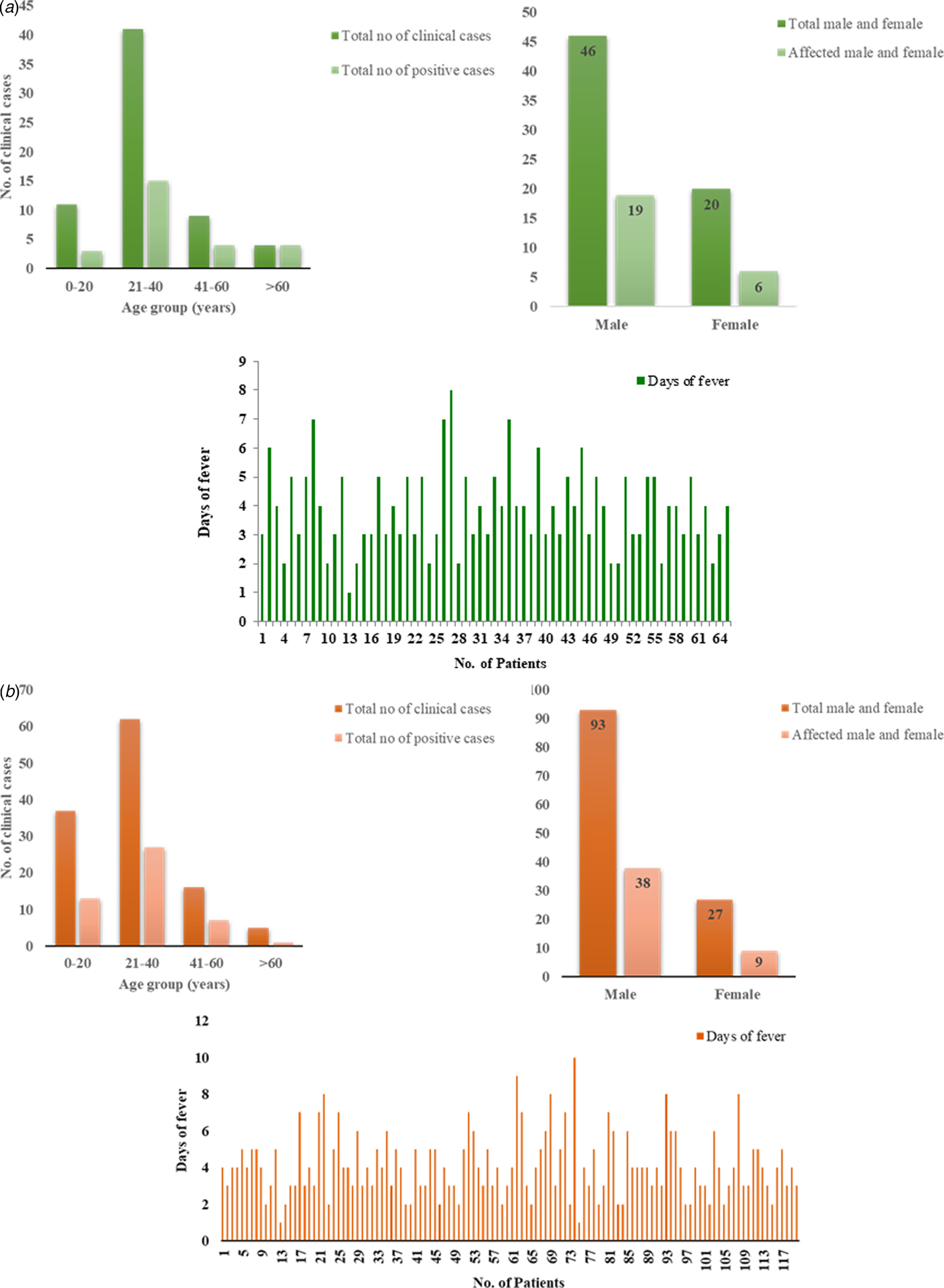
Fig. 2. Demographic details of the (a) CHIKV and (b) DENV patients. (i) The column graph showing correlation between number of clinical samples and CHIKV/DENV positive cases with the age group. (ii) The graph showing correlation between total percentage of males and females with affected cases. (iii) The graph showing total cases with days of fever.
Table 3. Correlation of demographic characteristics with CHIKV cases

Similarly, of the symptomatic DENV patients (n = 120), 92 (76%) were males and 28 (23%) were females with a male to female ratio of 3.2:1. The mean age of the symptomatic DENV patients was 28 years (s.d. ± 13.8) and duration of the acute phase of illness ranged from 1 to 10 days with mean 4 days (s.d. ± 1.74). The male to female ratio among the DENV virus-positive samples was 4.22:1. The mean age for the positive patients (48) was 24.81 years (s.d. ± 11.2), mean duration of illness was 3.6 days (s.d. ± 0.13). The age-wise analysis of the data suggested that maximum number of positive patients (27) belonged to 21–40 years age group followed by 13 patients in 0–20, seven patients in 41–60 and one patient in >61 age group (Fig. 2b, Tables 2 and 4). Five patients showed concurrent infection with CHIKV and DENV. The mean age and fever days of these patients were 17.4 years (s.d.SD ± 8.17) and 4.2 days (s.d.SD ± 1.30) respectively.
Table 4. Correlation of demographic characteristics with DENV cases

Two symptomatic patients had a low platelet count (0.7–08 lac/mm3). One of these patients (ID 36/16) was a 32 years male with four days of fever and platelet count of 0.7 lac/mm3. The other patient (ID 87/16) was 21 years male with four days of fever and platelet count of 0.8 lac/mm3. He had a low hemoglobin of 11.6. Both the patients were positive for DENV but negative for CHIKV. Three patients manifested slightly severe disease with mild hemorrhagic tendencies. All three patients were positive for DENV but negative for CHIKV.
DNA sequencing and accession numbers
A total of 19 study strains (11 CHIKV and 8 DENV) were sequenced for the investigation. The study strains were sequenced for the partial envelope protein gene (E1) for CHIKV and CprM region for DENV. The study sequences were confirmed by BLAST. The sequences were deposited in GenBank under the Accession Numbers for DENV-3 (KY099619–KY099620, MF038783–MF038788) and for CHIKV (MF774598–MF774608).
Molecular characterisation of CHIKV and DENV strains
Nine study CHIKV strains were used for the analysis. The nucleotide sequences of other two CHIKV strains were smaller in length (500 bp) and therefore were not utilised for the phylogenetic analysis. The nucleotide sequences of the study CHIKV strains (567 bp) corresponded to nucleotide position 10 555–11 121 of the genome of the prototype strain. The prototype strain of CHIIKV virus was S-27 (GenBank accession no. AF369024). A total of 67 CHIKV sequences of the E1 gene (including nine study sequences) were used for generation of phylogenetic tree. This analysis revealed that all the study strains clustered with ECSA genotype and grouped with other South Indian and South Asian sequences (Fig. 3a).

Fig. 3. (a) Neighbour-Joining Phylogenetic tree of CHIKV based on E1 gene and (b) Maximum Likelihood Phylogenetic Tree of DENV-3 based on CprM region. The study sequences are marked by the solid diamond (♦). The tree was constructed based on the Tamura-Nei model. Bootstrap values are represented by the numbers on nodes generated by 1000 replications. The study sequences clustered with the ECSA and genotype III respectively.
Seventy-one sequences of DENV-3 were used to construct the phylogenetic tree which included eight study strains and 63 sequences from GenBank. The prototype strain used for DENV-3 was H87 strain (GenBank accession no. M93130). The aligned region was 240 bp (80 amino acid) with respect to prototype strain. Phylogenetic analysis of CprM sequences demonstrated that all DENV-3 strains from New Delhi 2016 clustered with genotype III and grouped with other sequences from Asia (Fig. 3b).
Mutational analysis
The study sequences of CHIKV and DENV-3 strains were analysed with respect to the prototype strains. Mutational analysis showed that there were 18 and five mutations at nucleotide and amino acid levels respectively, of the CHIKV strains. The amino acid mutations in CHIKV strains included K211E, M269V, D284E, I317V and V322A. Similarly, we identified 20 and four mutations at nucleotide and amino acid levels respectively, in the DENV-3 sequences. Further, amino acid substitutions in DENV-3 were I133F, V19A, R35K and S75V. The amino acid changes in CHIKV and DENV-3 amino acid sequences are described in (Tables 5 and 6).
Table 5. Nucleotide and amino acid changes in the E1 gene of CHIKV

Table 6. Nucleotide and amino acid changes in the CprM region of DENV-3 serotype

Selection pressure analysis
The sequences of CHIKV and DENV-3 strains were subjected to selection pressure analysis to identify individual codon sites under positive selection. Five different methods namely, SLAC, FEL, REL, IFEL and MEME were used for the analysis. The dataset (n = 67) including all the three genotypes of CHIKV was used for the analysis. The low value of dN/dS ratio (< 1) suggested neutral selection in this region of the CHIKV genome. Four sites (211, 296, 304 and 321) were identified to be under positive selection in CHIKV (Table 7). The amino acid at position 211 was positively selected by three different methods (SLAC, FEL and IFEL).
Table 7. Selection pressure analysis of E1-CHIKV genome using SLAC, IFEL and FEL nucleotide substitution methods

Selection pressure analysis of the DENV-3 sequences was done by analysing non-synonymous to synonymous substitutions per site in the CprM region. The dataset (n = 71) analysed comprised of all genotypes of DENV-3 along with the study sequences This analysis revealed low mean dN/dS ratio (< 1) indicating that the codon positions were relatively conserved in the CprM region of DENV-3. Two codon sites (15 and 63) were under positive selection by REL and MEME methods (Table 8).
Table 8. Selection pressure analysis of DENV-3 using SLAC, FEL, MEME and REL nucleotide substitution methods

Shannon entropy analysis
The Shannon entropy analysis of all the CHIKV and DENV-3 strains was evaluated to find out the variable site in the study sequences. An entropy score of 0.2 was used as the threshold for the selection of variable sites. A total of 13 different variable sites were identified in CHIKV at positions 211, 213, 225, 226, 269, 276, 284, 296, 304, 317, 321, 343 and 344. Of these six codons at position numbers 211, 225, 226, 269, 284 and 304 had high entropy score (>0.5). Similarly, five variable sites were identified in DENV-3 at positions 13, 19, 35, 65 and 82. Two of these codon positions at 35 and 82 had high entropy value (>0.5) (Fig. 4a and 4b).

Fig. 4. Shannon Entropy plot of (a) E1 gene of CHIKV and (b) CprM region of DENV. Shannon entropy was analysed at different amino acids residues using Bioedit 7.0 software. Threshold value of entropy was set at 0.2. Highest entropy value shows the maximum number of chance of variability/mutation at that position given in the protein sequence.
Discussion
CHIKV and DENV are the arthropod-borne viruses that cause epidemics in several tropical and sub-tropical countries of the world. The common vector mainly contributes towards co-circulation and co-infection of these pathogens in endemic regions. The true disease burden of CHIKV fever is not known since diagnosis of CHIKV is not regularly implemented in clinical settings especially in DENV endemic regions. There is an increasing novel trend of concurrent DENV and CHIKV infections in epidemic/endemic DENV regions. Interestingly, there are many DENV-epidemic regions which have not reported cases of CHIKV infection. But, there are no report of outbreaks of CHIKV fever in a particular region without DENV infection [Reference Massad29].
We report co-circulation of CHIKV and DENV in DENV endemic region of New Delhi during 2016 after a gap of 5 years. The samples were collected from the OPD of a local health centre of Jamia Millia Islamia University in South East Delhi that is located near the banks of Yamuna River. Both CHIKV and DENV were detected in equal proportions i.e 40% of the samples tested by RT-PCR. Our earlier studies identified DENV and CHIKV in this region in 49% and 29% of samples respectively, during 2011. A previous study from Delhi reported preliminary data on CHIKV/DENV co-circulation and co-infection in 2016 [Reference Kaur12]. The novelty of the present study is that we carried out the molecular characterisation of circulating CHIKV and DENV during 2016 that was not reported earlier. Many earlier studies reported co-circulation of DENV and CHIKV viruses from India including from West Bengal (2011), Madhya Pradesh (2011–2012), New Delhi (2006, 2009, 2011), Maharashtra (2013), Tamil Nadu (2010–2012), Odisha (2013) and Karnataka (2011–2013) (Fig. 5a). Many other geographical regions have also reported co-circulation with both the viruses including Philippines (2012–13), Tanzania (2013), Gabon (2007–10), Lao PDR (2013), Yemen (2012), Mexico (2015), Brazil (2015), Columbia (2016), Angola (2014) including others (Fig. 5b). References/sources for these reports are given in Supplementary Table S2 and S3.
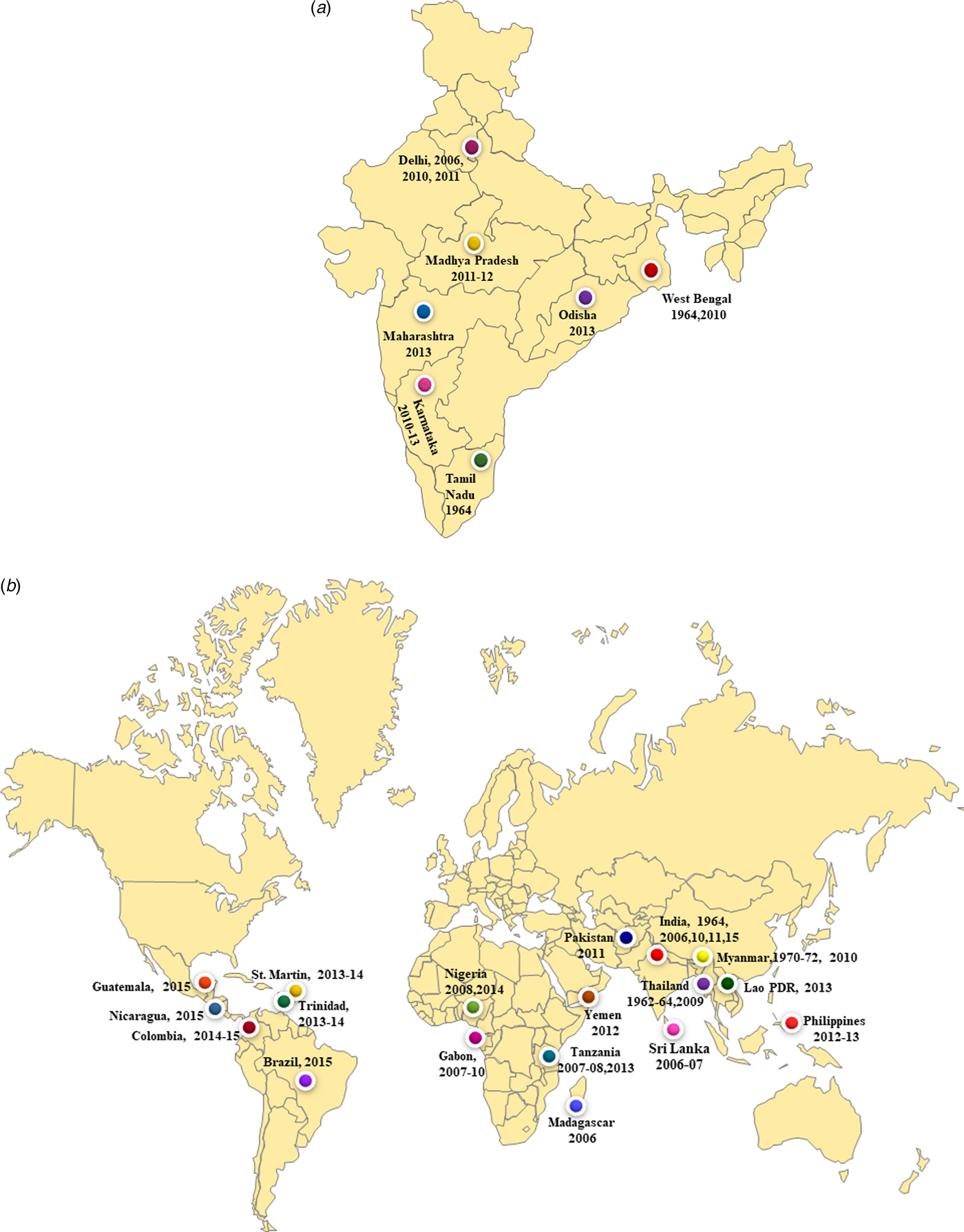
Fig. 5. (a) The co-circulation and co-infection of CHIKV and DENV in different parts of India. The freely available map of India was downloaded from the website presentationmagzine.com (https://www.presentationmagazine.com/powerpoint-map-of-india-647.htm) and edited in power point. (b) World map showing the co-circulation and co-infection of CHIKV and DENV in different geographical regions. The freely available map of India was downloaded from the website presentationmagzine.com (https://www.presentationmagazine.com/world-maps-vector-editable-507.htm) and edited in power point.
Co-infections with CHIKV and DENV were first reported from Vellore, Tamil Nadu, India in 1964 [Reference Myers and Carey30, Reference Carey31]. Another serologic study from southern India showed that these two viruses can co-exist in the same host [Reference Yergolkar32]. Co-infection was detected in 9% of the positive samples in the present study. The previous study from our laborataory also reported co-infection of both the virus in 10% of the sample in 2011 [Reference Afreen10]. Co-infection was also reported from New Delhi in 2006, 2010 with 8.7% cases. Other regions from India also reported concurrent infections with both the viruses in the range of 1–23%. The noteworthy co-infections from different parts of India included from West Bengal (12.4%), Odisha (13.7%) and Andhra Pradesh (23%). Many studies from other geographical regions also reported co-infection cases (0.9–18%). These include reports from Madagascar (18.2%), Tanzania (1%), Gabon (0.9%), Guatemala (10%), France (2.8%), Sri Lanka (5.5%). References/sources for these reports are given in Supplementary Table S2 and S3. Another interesting aspect of the investigation was concurrent infection with three different arthropod mediated pathogens (DENV, CHIKV and Plasmodium vivax) in two patients [Reference Tazeen27, Reference Abdullah28]. Such cases of infection with multiple pathogens pose a challenge for the diagnosis of the aetiological agents due to the overlapping clinical spectrum of the three diseases. Thus, the accurate and timely diagnosis of the pathogens is essential for effective patient management, especially in the endemic regions.
All the study strains of CHIKV clustered within ECSA genotype in the phylogenetic analysis. The study strains clustered with the strains reported from France (2010), Bhutan (2012), Australia (2008), Singapore (2006, 2008, 2009), Sri Lanka (2007, 2008), China (2010), Thailand (2009, 2010, 2013). Further, the analysis suggested that the study strains were closely related to the sequences described from India (HM159389) followed by Bhutan (KC731581) and France (FR846305). The DENV-3 strains clustered within the genotype III by the phylogenetic analysis. Genotype III has been previously reported from India [Reference Afreen33, Reference Tripathi34]. Phylogenetic analysis clustered the study strains with other recently reported sequences from India (2003, 2004, 2005, 2007 2008 and 2013) and adjoining countries including Pakistan (2006–2009) and China (2009).
CHIKV and DENV use humans and mosquitoes as the hosts for their propagation. The viral proteins have shown adaptation to the alternative hosts by the preference of synonymous mutations over non-synonymous ones. This process is known as negative or purifying selection. Selection pressure analysis of the E1 protein of CHIKV and CprM region of DENV-3 strains revealed strong purifying selection. The analysis revealed four and two sites in CHIKV and DENV respectively, under weak positive selection in the analysed regions. Weak positive selection pressure in these genomic regions has been reported previously [Reference Saswat35]. All the four amino acid positions in CHIKV showed positive selection and high entropy values suggesting probable variations at these sites. Further detailed mutagenesis studies on these amino-acid changes will define their role in the pathogenesis of CHIKV and DENV infections.
We were able to demonstrate simultaneous identification of CHIKV and DENV from New Delhi during 2016. Correlation of disease severity, genetic basis, immunity, climatic and environmental factors with simultaneous occurrence of these viruses will further augment delineation of the evolutionary trajectories of these viral pathogens. Further, in-depth investigation of the molecular and cellular pathways of the co-circulating viruses and their effect on the host immune response will provide more information on their co-evolution or otherwise. Simultaneous community and hospital-based longitudinal surveillance are needed for both the viruses for better patient management. This data will also provide intellectual input on the epidemiology of simultaneous occurrence of these viral infections, especially in the DENV endemic regions.
Supplementary material
The supplementary material for this article can be found at https://doi.org/10.1017/S0950268818001590.
Acknowledgements
We thank Indian Council of Medical Research (ICMR) to provide SRF-fellowship to Malik Hisamuddin for the present study. The study was supported by the King Saud University, Deanship of Scientific Research, College of Science Research Centre. This research received no specific grant from any funding agency, commercial or not for profit sectors.
Ethical statement
The authors assert that all procedures contributing to this work comply with the ethical standards of the relevant national and institutional committees on human experimentation and with the Helsinki Declaration of 1975, as revised in 2008.
Conflict of interest
None
















