Leber hereditary optic neuropathy (LHON) is one of the most common inherited optic neuropathy caused by mutations in mitochondrial DNA (mtDNA) that exhibit high phenotypic variability. Testing for primary LHON mutations (ND1/G3460A, ND4/G11778A, and ND6/T14484C) can greatly facilitate molecular diagnosis, identification of inheritance pattern, and disease-risk assessment.Reference Mackey, Oostra and Rosenberg 1 These mutations frequently occur with mitochondrial J1 haplogroup that probably acts as a background risk factor for the disease expression.Reference Brown, Starikovskaya and Derbeneva 2 Here we report a unique LHON patient who harbors a secondary LHON mutation (m.10663T>C) in the ND4L gene on a L3f1b (L3) haplogroup background.
Methods
The patient was enrolled at our neuro-ophthalmology clinic and a detailed clinical examination was performed as previously described.Reference Abu-Amero and Bosley 3 The study was approved by the Institutional Review Board and blood was drawn after obtaining informed consent. Mitochondrial DNA mutational analysis was done by direct Sanger’s sequencing as detailed elsewhere.Reference Abu-Amero and Bosley 3
Results
A 28 years old Saudi male presented with sub-acute painless loss of vision in his left eye which started three months prior to his clinic visit, with no other neurological symptoms. The patient had had a similar attack in his right eye a few months prior, with a diagnosis of central retinal vein occlusion. He had received an intravitreal injection of Bevacizumab with no visual recovery. Visual acuity in both eyes was 20/400 with normal intraocular pressure. Color vision was 0/15 on the right eye and 2/15 on the left eye. Pupil showed no relative defect and normal anterior segment.
Dilated fundus examination revealed diffuse disc paler with loss of papillomacular pundel in the right eye (Figure 1A) and pseudoedematous hyperemic disc with tortuosity of retinal arterioles in the left (Figure 1B). Visual field showed cecocentral scotoma in both eyes and denser inferiorly not respecting the midline with superior extension in the left eye (Figure 2).
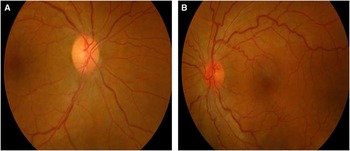
Figure 1 A: Mild diffuse paler with loss of papillomacular fibres in the right eye; B: Pseudoedematous hyperemic disc with tortuosity of retinal arterioles in the left eye
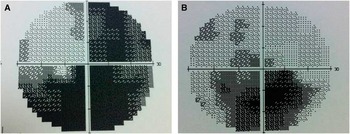
Figure 2 Visual field test showing cecontral scotoma with inferior extension
Mitochondiral DNA sequencing revealed a virtually homoplasmic m.10663T>C missense mutation in the proband, his mother and siblings had the mutation occurring at codon 65 in the transmembrane domain of ND4L gene that replaces a highly conserved valine to alanine (neutral AA). The change was absent in 235 healthy ethnicity-matched controls. PolyPhen2 (http://genetics.bwh.harvard.edu/pph2/bgi.shtml) and SIFT (http://sift.jcvi.org/) predicted this change to be ‘probably damaging’ and ‘affecting protein function’, respectively. As shown in the family pedigree (Figure 3) the proband is the only affected member in this family. His parents are unrelated and the mother was normal by examination. The proband is a non-smoker and reported no known maternal family history of LHON.
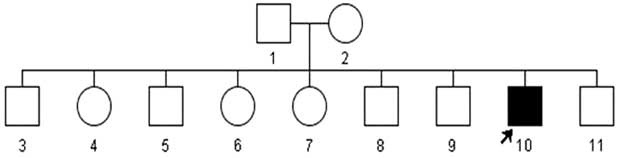
Figure 3 Family Pedigree of the proband. The arrow indicates the affected proband.
Sequencing of HVS-I and HVS-II regions showed the presence of L3f1b (L3) mtDNA haplogroup.
Discussion
The three primary LHON mutations are supposedly responsible for 90% of the LHON cases. However, most of these studies involved multi-generational families and patients with strong family history of LHON.Reference Mackey, Oostra and Rosenberg 1 In contrast, our previous study showed that only 17% (6/35) Saudi LHON patients had one of these primary mutations, wherein a complete mtDNA sequencing detected mutations in almost all the remaining patients.Reference Abu-Amero and Bosley 3 This indicated that clinically diagnosed LHON patients lacking the primary LHON mutations should be screened throughout the entire mitochondrial genome to detect other mutation(s) and highlighted the importance of other mtDNA mutations in LHON.Reference Abu-Amero and Bosley 3
Here we describe a previously reported secondary LHON mtDNA mutation (m.10663T>C) in the NDL4 gene of a Saudi LHON patient.Reference Brown, Starikovskaya and Derbeneva 2 The three primary LHON mutations (ND1/G3460A, ND4/G11778A, and ND6/T14484C) generally exist on a J1 mtDNA haplogroup. Similarly, the ND4L/T10663C also exits on a J1 mtDNA haplogroup as was observed in two cases of our previous study.Reference Abu-Amero and Bosley 3 However, in this study the mutation was present on a L3f1b mtDNA haplogroup. Mitochondrial haplogroup L3 has played a pivotal role in the history of the human species. Soon after the haplogroup arose in East Africa 84,000 and 104,000 years ago a relatively small number of migrants carried it across the Red Sea to Arabia, inaugurating an intercontinental migration that eventually settled every major land mass on Earth except Antarctica. That small group also gave rise to every non-African haplogroupReference Gonder, Mortensen and Reed 4 In a very recent study, Behbehani and colleagues described a large multi-generational pedigree from Kuwait with 14 LHON-affected members who did not have the primary mtDNA mutations but exhibited two uniquely concurrent mutations (10609T>C and 10663T>C) in the ND4L gene with a L3 haplotype.Reference Behbehani, Melhem and Alghanim 5 The patient reported in this study also seems to be a unique presentation with two plausible outcomes. First, m.10663T>C may be important to LHON without mtDNA background relevance. Second, both L3 and J1 may be important mtDNA haplogroups for LHON.
Conclusion
This report add to the importance of the m.10663T>C as a causative factor for LHON, possibly independent of the mtDNA haplogroup.
Acknowledgements
The study was supported by the Deanship of Scientific Research, Glaucoma Research Chair at the Department of Ophthalmology, College of Medicine, King Saud University, Kingdom of Saudi Arabia.
Disclosures
Majed Al-Kharashi, Abdullah Al-Kharashi, Majed Al-Obailan, Altaf A. Kondkar, and Khaled K. Abu-Amero do not have anything to disclose.





