On 26–27 May 2011, the 6th International Yakult Symposium was held in Vienna. The conference title (The Gut and its Role in Health Maintenance) reflected its objectives: to present and discuss the latest developments in understanding the complex relationship between the gut, its intestinal microbiota and health. The present study summarises key insights and learnings from the conference.
The opening keynote lecture from Professor Herbert Lochs (Innsbruck Medical University, Austria) gave a broad introduction to the topic, emphasising that one main function of the gut is to act as a barrier and defence against pathogens, allergens and harmful substances. The gut is of central importance for the body and overall health: for instance, the gut has a surface area of 300 m2; uses 40 % of the body's energy expenditure; contains 108 neurons and 50 % of the body's immune cells. New molecular analytical techniques are underlining the fact that the gut is home to diverse microbiota comprising about 1014 bacteria, representing up to 15 000 different species(Reference Frank, St Amand and Feldman1). The interactions between the host and its microbiota are key to overall health (Fig. 1). Mutually influencing interactions between the host genome, its resident microbiome, and physiological and environmental factors such as the diet are important in sustaining a well-functioning protective response and beneficial metabolic pathways in the gut.
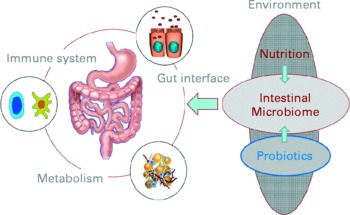
Fig. 1 The interactions of nutritional factors and the intestinal microbiome on gut-associated metabolic activities, barrier function and the immune system (Reproduced by permission of Professor Dirk Haller, Technische Universität München, Germany).
Health maintenance
How should health be defined and measured?
Not only is health difficult to define, but it is also difficult to measure, particularly when assessing health improvement or reduction of disease risk in apparently healthy individuals. Professor Renger Witkamp (Wageningen University, The Netherlands) stressed the important differences between nutritional products and pharmaceutical preparations in terms of application (healthy people v. diseased), biological activity and time dependence of effects, risk:benefit ratios and regulatory framework.
In Europe, the necessity for the approval of health claims for functional foods (Regulation (EC) No 1924/2006 of the European Parliament and of the Council on nutrition and health claims made on foods) by the European Food Safety Authority requires the scientific demonstration of beneficial physiological effect(s) in representative target groups. While clinical symptoms that can be markers of poor gut health and/or function are available, there are few, if any, biomarkers and/or clear parameters that are both validated and relevant for health maintenance for claims relating to gut and immune function. While the European Union (EU) regulations now also allow claims relating to the reduction of a ‘risk factor’ for a disease, food claims need a different approach from that used for pharmaceuticals, where effects on clinical or surrogate endpoints are easier to establish (Fig. 2).

Fig. 2 The differences between quantifying health and disease: pharmacological and disease biomarkers are not suitable for defining health. (Reproduced by permission of Professor R Witkamp, Wageningen University, The Netherlands, adapted from van Ommen et al. (2008) Genes Nutrition 3, 51–59).
A good starting point would be to have scientific agreement on a better working definition for health, based less on the WHO concept relating to the well-being and absence of disease(2, 3), and more on the ability of a person to adapt to internal and external stimuli in order to limit the loss of homeostasis. Professor Witkamp illustrated the continuum and gradual transition between health and a subclinical disease state. Closer examination of an apparently healthy individual might reveal an increased risk of disease due to an aberrant metabolic state. This may be indicated by subtle changes in a range of biomarkers and/or clinical conditions, for example by slightly elevated blood pressure, slightly reduced insulin response, slightly abnormal lipids, mild liver damage and elevated mediators of inflammation. Nutrition has an important influence on the metabolic state of an individual and thus the balance between homeostatic mechanisms. Dynamic processes are more useful indicators of health and disease risk than single endpoints, and models are now available that could achieve this. Genomics and systems biology can measure multiple changes in biomarkers, which can be translated into processes indicating health status or risk, e.g. low-grade inflammation, metabolism, oxidative stress, vascular function, stress responses, intestinal permeability, composition of the intestinal microbiota.
Measuring the robustness (or resilience) of physiological homeostasis in individuals was recommended as a promising approach, i.e. examining whether/how a person can revert to ‘normality’ or to modulate a disease risk factor after challenge with some sort of stress. As beneficial physiological effects are difficult to study in healthy people, challenge models and investigation of subjects with sub optimal health or risk of certain diseases would be appropriate. Examples of stress models are oral glucose and lipid tolerance tests, organ function tests, exercise or even psychological stress challenges.
For claims relevant to the resilience of intestinal health, measurements of both the dynamics of the intestinal flora and changes in gut barrier function are meaningful, taking advantage of novel techniques such as microarray analysis and metabolomics. A cross-over design might be preferable, using placebo where possible, so that each subject acts as their own control; this would also reduce inter-individual variation and the number of recruits required. Certain issues remain unresolved, however, including the precise nature of the challenge, the force of the stimulus, accepted designs, statistics and validation.
To understand the whole, one must study the whole
Professor Liping Zhao (Shangha Jiao Tong University, China) introduced the concept of a holistic approach to the interaction of food, microbiota and health. In Western countries, food is usually evaluated purely in terms of nutritional content but in China, where food and drugs are believed to derive from the same source, other properties are considered important. There is a tradition of using foods to prevent or combat chronic disease, with benefits demonstrated over thousands of years of ‘human trials’. Importantly, traditional Chinese medicine assesses and intervenes at the whole body level, using a holistic, dynamic and personalised approach. This reflects the modern concept of whole-body systems biology. Professor Zhao advised that ‘emergent functions’ of the body, e.g. its metabonome, metagenome and secretome, should be evaluated(Reference Volponi, Broteron and Luppold4). He stressed the importance of the intestinal microbiome to overall health, suggesting that in the future, health might even be monitored or predicted using intestinal microbiota-targeted, whole-body systems biology approaches(Reference Zhao and Shen5).
The intestinal epithelial barrier function and its protective effects
Professor Jörg-Dieter Schulzke (Charité Campus Benjamin Franklin, Berlin, Germany) described the key features of the epithelial barrier: an apical enterocyte membrane, an epithelial tight junction and an epithelial repair mechanism. The tight junctions, comprising (so far) twenty-seven different claudins, are the conduit for the transport of ions, water and macromolecules. If the tight junctions become disrupted, however, intestinal barrier dysfunction can ensue, for instance as induction of epithelial apoptosis or occurrence of epithelial ulcers and erosions (Fig. 3). The integrity of the barrier is important in many ways, with two examples given: (1) prevention of diarrhoea, which would result if ions and water are allowed to leak from the circulation into the intestinal lumen and (2) the concept of the ‘leaky gut’, where there is undesired uptake of antigens and bacteria from the intestinal lumen, which can be a trigger for inflammation.

Fig. 3 Causes and consequences of intestinal barrier dysfunction. (Reproduced by permission of Professor J.D. Schulzke, Technische Universität München, Germany).
Preserving the integrity of the epithelial barrier and preventing its dysfunction in inflammatory disease are research interests of Professor Schulzke's group. Anti-inflammatory remedies have been investigated, for example, in a study using TNF-α antibody therapy for 14 d with patients in active Crohn's disease(Reference Zeissig, Bojarski and Buergel6). The antibody therapy decreased and even normalised the raised epithelial apoptopic ratio, as well as improved the epithelial resistance. Recent research, presented as a poster at the symposium, showed that transforming growth factor-β exerts its protective effect on the intestinal mucosa by up-regulating the tight junction protein claudin-4 via Smad-4-dependent and independent transcriptional regulation(Reference Hering, Fromm and van Tol7). Other studies searching for treatments with direct barrier influence have investigated SCFA, flavonoids, growth factors, phytotherapeutics such as berberine, and phytotherapeutic. Butyrate, for example, appears to increase epithelial resistance; an effect linked to a decrease in claudin-2 mRNA. Positive effects of the flavonoid quercetin have been demonstrated in vitro using Caco-2 cells, which seemed to be linked to the up-regulation of the tight junction protein claudin-4(Reference Amasheh, Schlicter and Amasheh8). The plant alkaloid berberine, used in traditional Eastern medicine to treat diarrhoea, was shown to protect against TNF-α-mediated barrier defects in a human colon cell model and in a rat colon, by preventing the TNF-α-induced claudin-1 disassembly and the up-regulation of claudin-2(Reference Amasheh, Fromm and Krug9). Finally, some preliminary research with the probiotic Escherichia coli Nissle was shown: both live cells and supernatant increased epithelial resistance using the HT-29/B6 cell model. A poster at the symposium by Menz et al. (University of Tubingen, Germany) described a pre-clinical model of acute colitis which identified that this probiotic strain ameliorates symptoms via flagellin and the secreted protein tcpC.
The intestinal microbiota
A complex community: new findings from whole-systems analyses
Recent advances in genome sequence technology, high-throughput genomics data and comparative metagenomics have revolutionised microbiological research. The new approach enables the characterisation of entire microbial ecosystems, including unculturable species(Reference Raes and Bork10). Professor Jeroen Raes (Vrije Universiteit Brussel, Belgium) explained how these techniques allow a microbial snapshot to be taken. The Human Microbiome Project, launched in 2008, is aimed at characterising microbial communities at different sites in the body including the gastrointestinal tract, and analysing the role of these microbes in human health and disease(Reference Turnbaugh, Ley and Hamady11). The MetaHIT consortium, funded by the European Commission, is part of this project. The MetaHIT partners analysed faecal samples of 124 European individuals, and 3·3 million non-redundant microbial genes were Illumina-based metagenomic sequenced, assembled and characterised. The gene set, which was approximately × 150 that of the human hosts, was almost entirely of bacterial origin and contained up to 1500 prevalent bacterial species. Approximately one-third of these species was present in all individuals with 38 % of the gene pool shared by at least 50 % of the individual's samples(Reference Qin, Li and Raes12). A recent ground-breaking paper in Nature announced that, after combining twenty-two newly sequenced faecal metagenomes from subjects in European countries with previously published data, three robust clusters could be identified, which appear to be independent of the host's ethnicity or country(Reference Qin, Li and Raes12). These ‘enterotypes’ are stable constellations of co-occurring species with a main driver genus: Bacteroides (enterotype 1); Prevotella (enterotype 2); Ruminococcus (enterotype 3) (Fig. 4). Their biological impact remains unclear but preliminary data show that twelve genes significantly correlate with age, and three functional modules with BMI. Professor Raes speculated whether identification of an enterotype may help identify an individual's disease risk and allow customisation of their drug treatment. However, it is not yet known where these are, in fact, ‘enterotypes’ or ‘enterostates’, i.e. does the enterotype remain stable throughout life, does it change as a consequence of ageing, and can it be modulated, for instance with pre- or probiotics?

Fig. 4 The three enterotyopes of the human gut microbiome identified by the Metagenomics of the Human Intestinal Tract (MetaHIT) Consortium (see Arumugam et al.Reference Arumugam, Raes and Pelletier13 for full explanation). Newly sequenced faecal metagenomes from individuals from Denmark, France, Italy and Spain were compared with existing data from individuals from Japan and America. Visualisation of between class analysis of genus composition from different data sets (a – c) and (d) box plot showing the abundance of the main contributors of the three ‘enterotypes’ from one dataset. Reprinted by permission from MacMillan Publishers Ltd: NatureReference Arumugam, Raes and Pelletier13, copyright 2011. △, Obese; ![]() , IBD.
, IBD.
How does diet affect the intestinal microbiota?
Professor Alexander Haslberger (University of Vienna, Austria) emphasised the high level of metabolic activity of the intestinal microbiome, and how genetic factors, ageing, environment, pharmacological and chemical therapies, and diet constantly have an impact on the microbial profile. Data from different studies illustrate this: a less diverse microbiota and fewer Clostridium cluster IV (Ruminococcaceae) have been found in elderly people(Reference Zwielehner, Liszt and Handschur14); a vegetarian diet has been shown to affect the diversity of the Clostridium cluster XIVa(Reference Liszt, Zwielehner and Handschur15). A study by Professor Haslberger's group has also shown that chemotherapy combined with antibiotic treatment decreases absolute bacterial numbers, decreases Clostridium clusters XIVa and IV diversity, reduces levels of Faecalibacterium spp. and Proteobacter, and increases levels of Enterococcus faecium. The diversity of the intestinal microbiota has been shown to vary in different parts of the world, for example, being much greater in rural African children compared with those from the EU(Reference Greer and O'Keefe16). Current concern is that the Western diet (in particular its high content of animal-derived nutrients, lack of complex carbohydrates, overuse of antibiotics and low rates/duration of breast-feeding) increases the inflammatory potential of the intestinal microbiota.
Professor Harry Flint's group at the Rowett Research Institute (Aberdeen, UK) has investigated the potential health benefits from dietary non-digestible carbohydrates, which can be fermented under the anaerobic conditions in the colon to yield SCFA and other metabolites. The type of non-digestible material influences not just the metabolites produced, but also the species composition of the colonic microbiota. Early molecular profiling studies showed that while the dominant members in the colon remain relatively stable, there are considerable inter-individual differences(Reference Zoetendal, Akkermans and de Vos17). Carefully controlled dietary trials have shown that changes in dietary carbohydrate intake affect the composition of particular bacterial species and groups: for example, a recent study where diets high in type 3 resistant starch stimulated two groups of amylolytic bacteria, one related to Roseburia spp. and the other to Ruminococcus bromii, in the majority of obese subjects(Reference Walker, Ince and Duncan18). These microbiota changes were rapid (within a few days) and reversible. Conversely, obese volunteers on slimming diets low in total carbohydrate showed a decrease in bifidobacteria and butyrate-producing relatives of Roseburia spp.(Reference Duncan, Belenguer and Holtrop19), which was accompanied by altered metabolite profiles, including reduced butyrate formation(Reference Russell, Gratz and Duncan20).
Bacterially produced fatty acids have important influences on the host including stimulation of host receptors influencing gut motility and immune responses, lipogenesis (acetate), glucogenesis (propionate), provision of energy for colonocytes, regulation of gene expression and apoptosis, protection against colorectal cancer and colitis. They can also be toxic at high concentrations. Butyrate is particularly important for supplying energy to the gut epithelium and regulating host cell responses. A recent study analysing butyryl-CoA-transferase:acetate CoA-transferase gene revealed that the four most prevalent operational taxonomic units belonged to Eubacterium rectale, Roseburia faecis, Eubacterium hallii and an unnamed cultured species(Reference Louis, Young and Holtri21). In a more recent study, faecal samples were analysed from healthy young adults who were omnivores or vegetarians, and elderly omnivores. The butyryl-CoA-transferase:acetate CoA-transferase gene was found at the lowest copy levels in the elderly and at the highest levels in the vegetarians. Clostridium cluster XIVa (one of the main butyrate-producing groups in the colon) was more abundant in the vegetarians than in the elderly, leading the researchers to conclude that the microbiota of the elderly provides less butyrate, which may contribute to the increased risk of degenerative disease observed in this age group(Reference Hippe, Zwielehner and Liszt22).
Microbial influence on obesity-related disease
The concept that the intestinal microbiota can be the origin of chronic disease dates back to the theories of Metchnikoff. A range of cytotoxins, genotoxins and immunotoxins have now been identified that have been linked to diseases such as autism, cancer, obesity and diabetes(Reference Sandler, Finegold and Bolte23–Reference Cani, Amar and Iglesias25). Research in Professor Zhao's laboratory using multidisciplinary approaches now links the functions of the human microbiome to host metabolic phenotypes(Reference Li, Wang and Zhang26). Work here focuses on the association between the composition of the gut microbiome with an individual's health phenotype, e.g. obesity.
Animal models show that long-term consumption of a high-fat diet overrides host genetics, resulting in severe obesity and insulin resistance. An unhealthy diet also helps turn the intestinal microbiota from friend to foe; a change linked to several aspects of the metabolic syndrome(Reference Cani and Delzenne27). Furthermore, gut dysbiosis (a condition of microbial imbalance) has been linked to the chronic, systemic, low grade inflammation associated with age-related diseases. Recent studies in mice identified changes in key bacterial groups relating to their consumption of a high-fat or normal diet. In animals on a high-fat diet, bifidobacteria were almost absent; sulphate-reducing, endotoxin-producing Desulfovibrionaceae were at higher levels in animals with impaired glucose tolerance(Reference Zhang, Zhang and Wang28). Work from Professor Zhao's laboratory also showed that a energy-restricted diet resulted in as much as 50 % increase in the maximum lifespan of mice; corresponding changes in key microbial phylotypes correlated with this. Microbiota changes were also observed when mice were switched from high-fat diets back to normal chow.
Could such findings have any beneficial application for humans? Professor Zhao took the unusual step to answer this by recruiting himself in a single-person human volunteer study. Photographs taken after 5 years show improved health: reductions in weight (20 kg), blood pressure, heart rate, TAG, cholesterol, associated with increased levels of faecal Faecalibacterium prausnitzii. An interesting anecdote but insufficient for any scientific conclusions. A larger study involving 123 volunteers investigated the potential metabolic syndrome benefits of an intestinal microbiota-targeted, dietary intervention lasting 23 weeks. Preliminary data showed subsequent changes of gut microbiota in these subjects that could be associated with the observed improvement of the metabolic syndrome (L Zhao et al., manuscript in preparation).
Microbiotal immunomodulation: its effect on health
Professor Kenya Honda (The University of Tokyo, Japan) focused on the influence of the intestinal microbiota on the development of the mucosal immune system. Recent research has given fresh insight into the mechanism of activities underlying the immunoregulatory role of the microbiota, and in particular its involvement with the effects of IL-17-producing cells (the helper T cells, Th17) and IL-10-producing regulatory T cells (Treg cells). These cells play critical roles in maintaining tolerance to self-antigens and in suppressing excessive and harmful immune responses. CD4+ T cells in the intestinal mucosa comprise significant numbers of these cells, and germ-free animal studies have demonstrated the importance of the microbiota in maintaining their abundance(Reference Atarashi, Nishimura and Shima29).
Key components of the microbiota involved in this regulatory capacity have recently been identified. Professor Honda gave an update on the work of his group. Segmented filamentous bacteria (Gram-positive, spore-forming bacteria most closely related to Clostridium species), for example, have been shown to induce Th17 cells in the lamina propria of mice, correlating with increased expression of genes associated with inflammation and anti-bacterial activity(Reference Ivanov, arashi and Manel30). The group then investigated whether there was any link between clostridia and the accumulation of colonic Treg cells. Using animal models, they found that the spore-forming component of the indigenous microbiota promoted accumulation of Treg cells in the colonic mucosa, particularly Clostridium clusters IV and XIVa. When mice were colonised by a defined mix of forty-six strains of clostridia, a robust accumulation of colonic Foxp3++Treg cells and levels of transforming growth factor-β were observed(Reference Atarashi, Tanoue and Shima31). The clostridia activated intestinal epithelial cells to produce transforming growth factor-β, resulting in accumulation of Treg cells, which appear to occur via induction of Helios-negative iTreg cells. The clostridia also induced IL-10 expression in the Treg cells (Fig. 5). Previous researchers have shown that Clostridium clusters IV and XIVa form a smaller proportion of the faecal microbiota in irritable bowel syndrome (IBS) subjects compared with healthy controls(Reference Frank, St Amand and Feldman1). Low levels of F. prausnitzii, another micro-organism recently identified as having anti-inflammatory involvement, correlates with disease activity in Crohn's disease patients(Reference Sokol, Pigneur and Watterlot32).
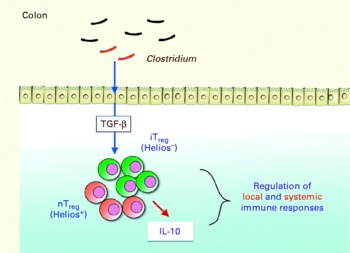
Fig. 5 Schematic indicating how Clostridium induces accumulation of regulatory T (Treg ) cells and IL-10 production in the colon, playing a critical role in the regulation of local and systemic immune responses. TGF-β, transforming growth factor-β; iTreg, inducible regulatory T cells; nTreg, naturally occurring regulatory T cells (Reproduced by permission of Professor K. Honda, The University of Tokyo, Japan).
Given the present understanding of the importance of the commensal biota in maintaining mucosal homeostasis and in triggering inflammatory diseases, it is crucial for future research to understand whether and how the indigenous microbiota affects the number and function of T cell subsets. The results presented here raise the possibility of therapeutic benefit by increasing the abundance of commensal species identified as important for the down-regulation of inflammation.
Microbiota involvement in functional gastrointestinal disorders
Professor Giovanni Barbara (University of Bologna, Italy) reminded the delegates of the impact of the IBS, in terms of sufferers' quality of life and the economic burden for healthcare. Many people are affected by this disorder, which has prompted interest in full elucidation of its pathophysiology. It is thought this involves a disturbance in the brain–gut axis but perhaps the brain–intestinal microbiota axis should also be considered. Not only can the brain affect the gut microbiota but conversely, the microbiota can affect behaviour(Reference Collins and Bercic33). Other lines of evidence further indicate intestinal microbiota involvement in IBS: the phenomenon of the post-infectious IBS(Reference Spiller and Garsed34), the microbiota's interaction with the motor apparatus of the gut(Reference Frank, St Amand and Feldman1, Reference Barbara, Stanghellini and Brandi35); its modulation of the hypothalamic–pituitary–adrenal system(Reference Sudo, Chida and Aiba36); and a recent study in rats(Reference O'Mahony, Marchesi and Scully37) that showed neonatal stress modifies the intestinal microbiota and evokes visceral hypersensitivity. Antibodies against flagellin, a component of the commensal flora, have been shown in certain IBS patients, suggesting an abnormal host-immune response towards components of the intestinal microbiota(Reference Schoepfer, Schaffer and Seibold-Schmid38). Compelling evidence also comes from demonstrations that modulation of the gut microbiota(Reference Preidis and Versalovic39) with probiotics(Reference Moayyedi, Ford and Talley40) and non-absorbable antibiotics(Reference Pimentel, Park and Mirocha41) can improve symptoms in certain patients.
The intestinal microbiota of IBS patients has often shown to be aberrant(Reference Malinen, Rinttilä and Kajander42–Reference Lyra, Rinttilä and Nikkilä44). This again raised the question of cause or effect: is an observed change in the intestinal microbiota in IBS patients a result of their altered gut function, or is the microbial change itself causing the disease symptoms? Could an ‘unhealthy’ intestinal microbiota abnormally stimulate the mucosal immune system through an excessively permeable mucosal barrier, and thus perturb bowel physiology and sensory perception to trigger IBS symptoms?
Most studies of IBS patients' microbiota have involved faecal analysis, initially with traditional culture-based techniques but now with high-throughput molecular-based techniques that allow a better phylogenetic characterisation. Recent studies have found, for example, lower levels of lactobacilli in diarrhoea-predominant IBS patients and higher levels of Veillonella spp. in those with predominant constipation(Reference Malinen, Rintatilä and Kajander45). Patients with the IBS have also been shown to have a higher concentration of bacteria in the mucosa compared with healthy controls. Breath tests can also be used to investigate the intestinal microbiota because certain gaseous metabolites are generated only by bacteria and not by the host(Reference Simrén and Stotzer46). Such a technique revealed that subsets of IBS patients may have small-bowel intestinal bacterial overgrowth, although this issue still remains controversial(Reference Lee and Pimentel47). Analysis of defensins, antimicrobial peptides produced by host Paneth cells in the intestinal crypts, indicates further negative host–microbial interaction: IBS patients have been shown to have higher expression of human β-defensin-2(Reference Salzman, Underwood and Bevins48, Reference Langhorst, Junge and Rueffer49).
An increase in gut permeability, resulting from disruption to tight junctions and leading to abnormal bacterial translocation, may underlie the mucosal immune activation observed in certain patients(Reference Turner50). In a recent study, a large proportion of IBS patients was found to show such increased mucosal permeability, which was associated with decreased gene expression of the tight junction structure zonula occludens(Reference Piche, Barbara and Aubert51). An increased infiltration of tryptase+mast cells and CD4+/CD8+ T cells along with a higher production of serine proteases, histamine and PG has been found in IBS, which may explain the disturbed sensory and motor function experience by these patients. Close vicinity of mast cells to mucosal nerves correlates with patients' pain severity and frequency(Reference Barbara, Stanghellini and De Giorgio52), and mediators released by the intestinal mucosa of patients resulted in an increased activation of sensory pathways, providing a functional link between immune activation and pain experience in IBS patients(Reference Barbara, Wang and Stanghellini53).
Probiotic research: an update in specific areas of health benefit
Probiotic research forms part of the evidence of the influence of the intestinal microbiota on health and disease. It is now widely accepted that regular ingestion of probiotics can modify the population of the gut microflora, thereby providing a practical means of enhancing or restoring gut and systemic immune function. Some of the presentations and posters focused on specific aspects of such research; a few key areas are reviewed here.
Mechanism of immunomodulation by commensal and probiotic micro-organisms
Professor Dirk Haller (Technical University of Munich, Germany) gave an update on current understanding into how probiotics influence the immune response, but he started by raising the same ‘chicken or egg’ question – is gut microbial dysbiosis a cause or effect of a disease state (Fig. 6)(Reference Spor, Koren and Ley54)? The answer to this is relevant to answering how and whether probiotic strains can ameliorate symptoms of disease and/or prevent disease onset. Meta-analyses indicate probiotic potential, to a greater or lesser extent, for the prevention and/or treatment of infectious and inflammatory conditions.

Fig. 6 Gut dysbiois associated with disease. Cause or effect? Microbial analysis of samples from patients and healthy controls showing relative abundance of predominant bacterial phyla. (a) Caecal samples and inflammatory bowel disease; (b) faecal samples and type 2 diabetes; (c) faecal samples and necrotising enterocolitis ![]() , Firmicutes;
, Firmicutes; ![]() , Bacteroidetes;
, Bacteroidetes; ![]() , Fusobacteria;
, Fusobacteria; ![]() , Actinobacteria;
, Actinobacteria; ![]() , Verrucomicrobia;
, Verrucomicrobia; ![]() , Proteobacteria. Reprinted by permission from Macmillan Publishers Ltd: Nature Reviews Microbiology, from Spor et al. (Reference Spor, Koren and Ley54), copyright 2011.
, Proteobacteria. Reprinted by permission from Macmillan Publishers Ltd: Nature Reviews Microbiology, from Spor et al. (Reference Spor, Koren and Ley54), copyright 2011.
The exact pathways involved in probiotic activity in targeting the gut barrier functions and regulating the immune response in the gut are being unravelled at the most basic level. Macromolecules on the surface of probiotic bacteria are important for strains to interact with host pattern recognition receptors on the gut mucosa(Reference Lebeer, Vanderleyden and De Keersmaecker55). Toll-like receptor signalling is important, resulting from recognition of bacteria and their products by the host and leading to epithelial cell proliferation, secretion of IgA into the gut lumen and expression of antimicrobial peptides(Reference Abreu56). Such pro-inflammatory signals help to maintain epithelial cell integrity and barrier function as well as induce immunoregulatory mechanisms that control adaptive immune functions. Professor Haller's group, working with both non-pathogenic Gram-negative enteric bacteria(Reference Haller, Russo and Sartor57) and colitogenic Enterococcus faecalis (Reference Ruiz, Shkoda and Kim58), has shown that bacterial signals can trigger transient activation of the pro-inflammatory transcription factor NF-κB. A proof-of-concept study in mice indicated that the intestinal epithelium is reactive towards environmental changes. Administration of a commensal Lactobacillus reuteri strain induced transient activation of the intestinal epithelial cells, even though the mice had already developed a complex microbiota(Reference Hoffmann, Rath and Holzwimmer59).
To maintain health and to avoid inflammation or loss of barrier integrity, it is important that there is the right balance of activation. The elucidation of inflammatory bowel disease (IBD) aetiology may benefit from data derived from microbial investigations(Reference Kaser, Zeissig and Blumberg60). A recent study with genetically susceptible IL-10-deficient mice, for example, revealed that a metalloprotease produced by commensal strains of E. faecalis contributed to the development of chronic colitis, by damaging the integrity of the epithelial barrier(Reference Steck, Hoffmann and Sava61).
Intestinal epithelial cells and dendritic cells interact with and respond via their pattern recognition receptors that detect microorganism-associated molecular patterns. Currently, it is not possible to delineate which bacteria surface molecules can be regarded as health-promoting (and therefore of probiotic relevance) rather than pathogenic. Only a limited number of microorganism-associated molecular patterns–pattern recognition receptors interactions of probiotics (and pathogens) are known(Reference Lebeer, Vanderleyden and De Keersmaecker62); identification of structure–function relationships for more strains will help substantiate probiotic mechanisms of activity.
Intestinal epithelial cells are at the interface between luminal (thus, including bacteria) and host-derived signals; any disruption of this communication negatively affects the intestinal barrier function, triggering mucosal immune disorders in susceptible people(Reference Clavel and Haller63). Recent research into Crohn's disease in a murine model by Professor Haller's group has found that a diet lacking in Fe sulphate prevented the onset of chronic ileitis and also resulted in substantial changes in the intestinal microbiota, for example by reducing the abundance of Desulfovibrio (Reference Werner, Wagner and Martinez64). The results suggested that luminal Fe promotes disease onset via changes in the microbial composition that trigger epithelial cell stress-associated apoptosis.
Probiotics and adjuvant effect
Professor Lorenzo Morelli (Istituto di Microbiologia UCSC, Piacenza, Italy) opened by commenting that the concept of modulation of the gut-associated lymphoid tissue by probiotics is well established and known to be strain-dependent. In fact, the first industrial application of immune modulation by lactic acid bacteria focused on their adjuvant activity(Reference Schiffrin, Rochat and Link-Amster65). Investigations of probiotic immunogenicity have focused on two lines of research: (1) the use of GM probiotic strains to deliver specific antigens into the gut; and (2) the use of unmodified probiotic strains to improve vaccine efficacy against different diseases (adjuvant effect).
Research interest in the adjuvant potential of probiotics will no doubt increase because of a recent reference made in the guidance from the European Food Safety Authority for scientific requirement for health claims relating to gut and immune function(66). This states that higher vaccination responses are beneficial, and stimulation of protective antibody titres, as measured by increased numbers of individuals attaining protective levels, could be used to substantiate immune claims relating to defence against pathogens.
To date, human studies have been published reporting beneficial probiotic adjuvant effects with viral vaccines (e.g. influenza and hepatitis) and vaccines comprising attenuated pathogenic bacteria (e.g. Salmonella, cholera, Pneumococcus, Haemophilus). For example, live-attenuated influenza vaccines have been used to show the adjuvant effects of probiotics in four studies with the elderly(Reference Bunout, Barrera and Hirsch67–Reference Davidson, Fiorino and Snydman70). The studies used broadly similar clinical protocols: probiotic consumption for a period of 1 month to 13 weeks, with administration of the vaccine sometime during this intervention. Some differences in the specific adjuvant action were observed, particularly with regard to the specific flu viruses. After reviewing all the available data, Professor Morelli came to a number of conclusions: lactobacilli look promising as mediators of the adjuvant effect, whereas bifidobacteria do not; probiotic adjuvant effects seem to be stronger with viral vaccines; viable probiotic cells are probably better than dead; and, finally, more research is needed.
In a study investigating the strain specificity of probiotic adjuvant effect, the immunomodulatory properties and sublingual immunotherapy capacity of eleven strains of lactic acid bacteria were compared(Reference Van Overtvelt, Moussu and Horiot71). This identified two groups: those that strongly induced IL-12p70 and IL-10 in dendritic cells, supporting interferon-γ and IL-10 production in CD4+ cells (e.g. L. helveticus), and those that were pure Th1 inducers (e.g. L. casei). Based on the effects of these strains in a murine asthma model, it was concluded that strains acting as Th1/possibly Treg inducers, but not Th1 adjuvant, were more suited as an adjuvant for sublingual allergy vaccines.
Selection of the best Lactobacillus strains for adjuvant benefit would be helped by focusing on their main aspects of immune interaction: the bacteria's surface proteins and adhesion properties, surface glycoproteins and secreted metabolites. A surface protein of interest is the S-layer, which is composed of protein monomers in regular arrays(Reference Callegari, Riboli and Sanders72). Using knockout mutants, the S-layer protein A of L. acidophilus NCFM has been shown to be involved in the regulation of immature dendritic cell and T cell functions(Reference Konstantinov, Smidt and de Vos73). Secreted substances, such as H2O2, may also be of interest(Reference Voltan, Martines and Elli74). Such research needs to be continued in order to fully exploit the potential of probiotics as adjuvants, and to gain clear scientific evidence of strain-specific beneficial immunomodulatory effects.
Probiotic research into colorectal cancer benefit
Professor Joseph Rafter (Karolinska Institutet, Stockholm, Sweden) stressed why probiotic research in this area is needed, by reminding delegates that colorectal cancer (CRC) is the third most common cancer in men and the second in women worldwide(Reference Boyle and Ferlay75). Lifestyle factors play a major role in CRC aetiology, i.e. the combination of four dietary factors (fibre, fish, red and processed meats) in addition to alcohol intake, obesity and low physical activity(76).
The colonic microbiota may be involved in the aetiology of this disease, thus its beneficial manipulation may have cancer-preventive effects. Currently, evidence is stronger for probiotic benefit(Reference Commane, Hughes and Shortt77) compared with prebiotics, but there are also some positive data with synbiotics. Data are, however, mostly derived from in vitro and animal models; human evidence, in terms of both epidemiological and interventional studies, is still insufficient.
To update on the current situation, Professor Rafter reviewed studies done in rats where good probiotic anti-genotoxic effects in the colon have been shown. As was noted in other areas of research, live cells seem to be needed and efficacy is species-dependent. Aberrant crypt foci, abnormal clusters of cells, are good early indicators of CRC development and have been used as research biomarkers. Pro-, pre- and synbiotic studies have shown aberrant crypt foci inhibition, influenced by the basal diet of animals(Reference Reddy78). Evidence for both pro- and prebiotics is also good in animal studies with regard to tumour prevention. Efficacy varies according to the genus or species of the probiotic strain, and effects were better if the probiotic was administered before the carcinogen.
Data from human epidemiological studies, while being varied and limited, are also difficult to interpret as most examined the effects of consuming fermented dairy products, which may not necessarily have been probiotic(Reference Kawano, Ishikawa and Nakamura79). In terms of human intervention studies, most have been done in healthy volunteers and investigated lactobacilli strains. Early studies explored a range of biomarkers, including faecal enzymes, urine/faecal mutagenicity, with results indicating that probiotics had the potential to reduce disease risk. Professor Rafter emphasised that, although approximately twenty to thirty biomarkers have been studied, most have not been validated against tumour development. He concluded that good CRC biomarkers indicating the risk, onset and development of this disease still need to be identified and validated.
Dietary intervention with two probiotic strains was shown to modulate the potential of human faecal water in inducing damage in a human colon tumour cell line(Reference Oberreuther-Moschner, Jahreis and Rechkemmer80). Only one study to date has examined probiotic effect directly on tumour development; in this study, the probiotic was shown to delay the progression of atypia of new tumours in patients at high risk of developing CRC (they had had prior surgical resection of CRC tumours)(Reference Ishikawa, Akedo and Otani81). Thus, the currently available data are insufficient to come to definitive conclusions on probiotic benefit.
Professor Rafter then described the SYNCAN project, a multi-partner study funded by the EU, investigating a synbiotic combination and cancer prevention. A full range of biomarkers and tests would be used throughout the project, including numerous colon mucosa biomarkers, various faecal water activities, and several immunological and inflammatory response markers in the colon and blood samples. The synbiotic combination chosen for study was a prebiotic enriched with oligofructose with two strains (L. rhamnosus and Bifidobacterium lactis). Initial long-term rat studies with this synbiotic found it to be protective against induced carcinogenesis. This protection appeared to be mediated by effects on SCFA, proliferation, glutathione-S-transferase P, inducible NO synthase and cyclo-oxygenase-2. The intervention primarily modulated IL-10 production and the cytotoxic activity of natural killer cells isolated from Peyer's patches(Reference Femia, Luceri and Dolara82). These results prompted a 12-week human intervention in polypectomised and colon cancer patients(Reference Rafter, Bennett and Caderni83). The synbiotic was associated with apparently beneficial changes in the intestinal microbiota: increasing lactobacilli and bifidobacteria while decreasing putrefactive groups such as Clostridium perfringens and coliforms. There was a corresponding reduction of colorectal proliferation in polyp patients. All the relevant biomarkers indicated cancer-protective potential in reducing exposure to genotoxins in the gut. As only minor effects were observed for systemic immune markers, it was assumed that the gut-associated lymphoid tissue might be more affected.
While there are indications for pro-, pre- and synbiotic benefits for the prevention of CRC, more human research is needed and many questions still need to be answered. For example, a range of mechanisms of activity are probably involved, but do different strains and species act by similar mechanisms? Is survival in the gut a prerequisite for efficacy? Dose and time response data are also currently lacking. Results from animals need to be corroborated in human studies, preferably using strains identified as having good anti-tumour effects, validated biomarkers and subjects with a high risk of the disease. Epidemiological studies need to be specific for probiotics, not just foods fermented with lactic acid bacteria. Professor Rafter finished by explaining the ongoing EU-funded TORNADO project, which is using a mechanistic approach to identify novel molecular targets for functional foods (including probiotics) and to investigate the effects of ageing on the intestinal microbiota. Importantly, this project could help industry by identifying new biomarkers for monitoring health (not just disease) and perhaps even provide a ‘template protocol’ that could be used for the substantiation of health claims.
Probiotic research with athletes
Professor Michael Gleeson (Loughborough University, UK) began by explaining how transient depression of the immune function can be caused by the prolonged intense exercise undertaken by high-performance athletes during their training and competition. This, for example, was demonstrated by a study conducted in competitive sailors, where a clear correlation was found between increased training/competition load and decreased levels of salivary IgA(Reference Neville, Gleeson and Folland84). Exercise-induced immune suppression is a major factor behind athletes' increased susceptibility to upper respiratory tract infections (URTI)(Reference Gleeson85), as was demonstrated in a study of 2311 runners who had a higher incidence of URTI during the week after they had taken part in the 1987 Olympic marathon competition, compared with a control group(Reference Nieman, Johanssen and Lee86). Athletes are more prone to URTI not just because of the physical stress of exercise; factors such as their increased psychological stress combined with possibly inadequate diet, foreign travel across time zones, disturbed sleep, exposure to environmental extremes, etc. can also result in a degree of immune depression that is probably additive to the effects of intensive exercise. Their exposure to pathogens may also be increased because of elevated lung ventilation during exercise, skin abrasions and exposure to large crowds. Some athletes are also susceptible to the development of gastrointestinal symptoms (e.g. abdominal discomfort, diarrhoea), which may particularly occur during long-distance runs or competition(Reference Simons and Kennedy87). These symptoms may not necessarily be due to infection; gut ischaemia-associated leakage of bacterial endotoxins into the circulation may be the cause(Reference Jeukendrup, Vet-Joop and Sturk88).
The current understanding of the range of probiotics' mechanisms of activity and their immune and gut benefit has prompted research into their potential for athletes. Athletes are also good subjects for challenge models to obtain evidence for health claims. Professor Gleeson reviewed the various intervention studies that have been conducted in athletes with probiotics. In these studies, a range of efficacy has been reported that may be due to strain-specific effects. A trial in marathon runners with L. rhamnosus GG found no difference in the incidence of URTI or gastrointestinal episodes(Reference Kekkonen, Vasankari and Vuorimaa89), but a trial with L. casei DN-114001 in army cadets during 3 weeks of combat training was probably too short to show any effect on URTI but did show that the probiotic was associated with improved maintenance of salivary IgA levels(Reference Tiollier, Chennaoui and Gomez-Merino90). A trial conducted in Australia in elite healthy male distance runners with L. fermentum VRI-003 was able to show that consumption of this strain was associated with improved health: the runners on probiotic experienced substantially fewer days and less severity of respiratory illness(Reference Cox, Pyne and Saunders91).
In a double-blind, placebo-controlled study in endurance athletes based at the Loughborough University, the effects of ingesting L. casei Shirota were investigated during a 4-month period of winter training and competition. A range of benefits were shown to be associated with probiotic consumption, including a lower proportion of subjects experiencing one or more weeks with URTI symptoms, a lower average number of URTI episodes, as well as a lower proportion of days that subjects suffered gastrointestinal symptoms(Reference Gleeson, Bishop and Oliveira92). These effects may have partly been due to salivary IgA concentrations, which were higher in the probiotic group compared with the placebo group. The strain specificity of any such effect was highlighted in a similar study using another Lactobacillus probiotic strain (L. salivarius), which resulted in no beneficial effects. He finished by emphasising the need for larger-scale trials to confirm efficacy but stressed the potential benefit of certain probiotics for athletes, particularly if they are travelling abroad or prone to illness.
Conclusions
In light of the fundamental and underlying purpose for probiotic use, namely for health maintenance, there is a clear need to revisit how health is defined and measured. A better approach could be based on the ability of a person to adapt to internal and external stimuli in order to limit the loss of homeostasis.
Substantiation of probiotic claims for gut and immune health benefits is hampered by the current lack of relevant and validated biomarkers. New ‘omic’ technologies may in future enable health to be monitored or even predicted using intestinal microbiota-targeted, whole-body systems biology approaches. The recent discovery of three enterotypes of the human gut microbiome, independent of the host's ethnicity or country, is an important advance in understanding. The biological impact of these enterotypes needs to be investigated further, but it was suggested that identification of an individual's enterotype may help identify their disease risk and allow customisation of their drug treatment and/or probiotic intake.
A combination of cultural and molecular studies is also delivering new insights into the impact of different diets on the individual variation in microbiota composition and subsequently upon their health outcome. The type of non-digestible carbohydrates ingested influences not just the metabolites produced but also the species composition of the colonic microbiota. Current concern is that the Western diet (in particular its high content of animal-derived nutrients, lack of complex carbohydrates, overuse of antibiotics and low rates/duration of breast-feeding) increases the inflammatory potential of the intestinal microbiota. Weight loss in overweight individuals has shown a change to an apparently much healthier phenotype with a reduced risk of disease, associated with microbiotal changes.
Gut dysbiosis has been linked to the chronic, systemic, low-grade inflammation associated with age-related diseases. These inflammatory processes are, among other factors, determined by the integrity of the gut epithelial barrier. Preliminary research has been presented showing that the probiotic strain E. coli Nissle, as live cells as well as supernatant, increased epithelial resistance and beneficial effects in a preclinical model of acute colitis. Given the present understanding of the importance of the commensal biota in maintaining mucosal homeostasis and in triggering inflammatory diseases, future research should focus on understanding whether and how the indigenous microbiota affects the number and function of T cell subsets. Results presented during the conference raise the possibility of therapeutic benefit by increasing the abundance of commensal species identified as important for the down-regulation of inflammation. Several lines of evidence also indicate intestinal microbiota involvement in IBS. A new perspective may be that the brain–intestinal microbiota axis is involved in the aetiology of the disease. An observed increase in gut permeability in IBS patients may underlie the mucosal immune activation.
In the sessions on specific areas of health benefit, the strain specificity of probiotics in enhancing vaccine efficacy was reviewed. It was concluded that lactobacilli, rather than bifidobacteria, look promising as mediators of the adjuvant effect, particularly if live cells are used and with viral vaccines. A second topic dealt with colorectal cancer; the colonic microbiota has been implicated in the aetiology of this disease, indicating that beneficial manipulation of this microbial population may have a preventive effect. As in many other areas of discussion, the need for validated biomarkers and further research was emphasised. Finally, the influence of lifestyle factors on the immune system was illustrated by trials in high-performance athletes where prolonged intense exercise caused transient depression of immune function. Probiotic benefits for elite athletes may also be strain-specific. Positive results were shown for L. casei Shirota, where maintenance of immune function was observed to be associated with a reduction of the episodes of respiratory and gastrointestinal tract infections.
Acknowledgements
The 6th International Yakult Symposium was financially supported by Yakult Europe B.V., Schutsluisweg 1, 1332 EN Almere, The Netherlands. The authors declare that there is no conflict of interest. This work was commissioned by Yakult Europe B.V., and written by Dr Thomas (Yakult UK Limited) and Dr Ockhuizen (a consultant for Yakult Europe B.V.).








