Intestinal dendritic cells (DC) are central in controlling immune tolerance in the gastrointestinal tract( Reference Bernardo, Sánchez and Al-Hassi 1 ). DC capture antigens based on the expression of a wide variety of pattern-recognition receptors, including Toll-like receptors (TLR), nucleotide-binding oligomerisation domain-like receptors and C-type lectin receptors( Reference Dzopalic, Rajkovic and Dragicevic 2 , Reference Bermudez-Brito, Plaza-Díaz and Muñoz-Quezada 3 ), which play crucial roles in host recognition of both probiotics and foreign micro-organisms( Reference Meijerink and Wells 4 ). These interactions induce signalling cascades and trigger a molecular response against the detected micro-organism, including the production of immunomodulatory cytokines and chemokines, antimicrobial or cytoprotective factors and co-stimulatory molecules( Reference Lebeer, Vanderleyden and De Keersmaecker 5 ). Bifidobacterium and Lactobacillus are the major commensal microbes of the gastrointestinal tract that are frequently used as probiotics( Reference Rizzello, Bonaccorsi and Dongarrà 6 ). In addition, recent reports have shown that probiotic treatment can modulate DC, as several Lactobacillus species regulated DC surface marker expression and cytokine production and maturation( Reference Kim, Ohta and Takahashi 7 – Reference Christensen, Frøkiaer and Pestka 9 ). However, the response of the immune system to probiotics remains controversial. Some probiotic strains modulate the production of cytokines by DC in vitro and induce a regulatory response, whereas others induce a pro-inflammatory response( Reference Evrard, Coudeyras and Dosgilbert 10 ). These strain-dependent effects are thought to be linked to specific interactions between bacteria and pattern-recognition receptors.
Escherichia coli is a common inhabitant of the human gut, where it acts as a harmless component of the normal flora. However, in some cases, it can be responsible for severe diarrhoea, especially in children( Reference Santona, Diaz and Fiori 11 ). Some studies have shown that probiotics have the potential to beneficially modulate the cytokine balance, promoting potent type-1 immune responses or preventing immune dysregulation associated with specific T-cell polarisation( Reference Bermudez-Brito, Plaza-Díaz and Fontana 12 ). For instance, probiotic bacteria alter the activation, expression and secretion of pro-inflammatory cytokines (such as TNF-α) and chemokines (such as chemokine (C–X–C motif) ligand-8) caused by enteric pathogens( Reference Gareau, Ho and Brenner 13 ). However, these studies did not determine the mechanism(s) responsible for these probiotic immunomodulatory effects. Therefore, the aim of the present study was to (1) investigate the ability of Lactobacillus rhamnosus CNCM I-4036 and its cell-free culture supernatant (CFS) to activate human intestinal DC, (2) determine how they respond to pathogenic bacteria, specifically E. coli, and (3) elucidate the molecular mechanisms involved in these interactions. L. rhamnosus CNCM I-4036 was isolated from the faeces of breast-fed newborn infants, as described in a previous study( Reference Vieites Fernández, Muñoz Quezada and Llamas Company 14 ), and selected based on its probiotic properties, such as adhesion to intestinal mucus, sensitivity to antibiotics and resistance to gastrointestinal juices, biliary salts, NaCl and low pH. The majority of studies that involved intestinal DC are based on mouse models, and research regarding human gut DC is limited, primarily due to methodological difficulties in studying human tissue and isolating human gut DC. Therefore, we used human DC that extend dendritic processes and that sample antigens in a manner similar to the way lamina propria DC sample luminal antigens.
Materials and methods
Ethics statement
The present study was approved by the University of Granada Ethical Committee. L. rhamnosus was obtained from the faeces of breast-fed newborn infants according to the protocol( Reference Vieites Fernández, Muñoz Quezada and Llamas Company 14 ).
Bacterial and supernatant preparation
The isolated probiotic strain L. rhamnosus CNCM I-4036( Reference Muñoz-Quezada, Chenoll and Vieites-Fernández 15 ) was cultured in De Man–Rogosa–Sharpe broth medium (Oxoid) for 18–24 h at 37°C under anaerobic conditions. The CFS was obtained by centrifugation at 12 000 g for 10 min, neutralised with 1 m–NaOH to a pH of 7·0, concentrated 10-fold by lyophilisation and then sterilised by filtering through a 0·22 μm filter (Minisart hydrophilic syringe filter; Sartorius Stedim Biotech GmbH). The supernatant was added at a concentration of 7 % (v/v).
The Spanish Type Culture Collection (CECT) provided the pathogenic strains. The pathogens used in the present study included E. coli CECT 742, CECT 515 and CECT 729. The cultures were propagated aerobically in tryptone soya broth (Panreac Química).
In a previous study, we investigated the ability of L. rhamnosus CNCM I-4036 and its supernatant to inhibit the growth of different E. coli strains by monitoring bacterial growth at 37°C in tryptone soya and analysing the optical density at 620 nm. Our data showed that live L. rhamnosus significantly inhibited the growth of enterotoxigenic E. coli (CECT 501 and CECT 515) and enteropathogenic E. coli (CECT 727 and CECT 729). In the presence of non-neutralised supernatant, L. rhamnosus CNCM I-4036 inhibited the growth of these enteropathogens by 10–50 %; however, upon neutralisation, these effects were diminished except against E. coli CECT 742 (data not shown). Based on our previous study and in order to avoid changes in the media and DC death, live L. rhamnosus were incubated with E. coli CECT 515 and 729, whereas its supernatant was incubated with E. coli 742.
Before the experiments, pathogens were cultured at 37°C in tryptone soya broth for 8 h and then subcultured at a dilution of 1:500 in Roswell Park Memorial Institute-1640 medium (Sigma-Aldrich) containing 10 % fetal bovine serum (Gibco Invitrogen) at 37°C overnight.
Cell preparation
DC generated from umbilical cord blood CD34+ progenitor cells (haematopoietic stem cells) were supplied by MatTek Corporation( Reference Ayehunie, Snell and Child 16 ). These cells were seeded onto twenty-four-well plates in DC maintenance media (MatTek).
Bacterial co-culture and dendritic cell stimulation
DC were stimulated as described previously( Reference Bermudez-Brito, Muñoz-Quezada and Gomez-Llorente 8 ). Briefly, DC were directly challenged for 4 h with live L. rhamnosus (107 colony-forming units/ml)/CFS, E. coli (106 colony-forming units/ml) or both. After incubation, the media were removed and replaced with fresh DC maintenance media. After 20 h, culture supernatants were collected for cytokine analysis, and the cells were collected for RNA extraction. E. coli lipopolysaccharide (20 ng/ml; Sigma-Aldrich) was used as a positive control. Negative control cultures contained unstimulated DC.
Cytokine and chemokine quantification in culture supernatants
The amount of cytokines present in the supernatant was measured using MILLIplexTM immunoassays (Linco Research, Inc.) using the Luminex 200 system according to the manufacturer's instructions. IL-1β, IL-6, IL-8, IL-10, IL-12p40, IL-12p70, TNF-α, interferon-γ, monocyte chemotactic protein 1/chemokine (C–C motif) ligand (CCL)-2, macrophage inflammatory protein-1α/CCL-3, RANTES (regulated upon activation, normal T-cell expressed and presumably secreted)/CCL-5, macrophage-derived chemokine/CCL-22 and transforming growth factor (TGF)-β were evaluated. Data are representative of three independent experiments.
RT reaction and PCR
Inflammatory response in the bacteria-treated DC was examined using Human TLR Signaling Pathway PCR Arrays, as described previously by Bermudez-Brito et al. ( Reference Bermudez-Brito, Muñoz-Quezada and Gomez-Llorente 8 ). Each array comprised primer pairs that are specific to twenty genes related to TLR-mediated signalling pathways.
Briefly, DC were lysed and total RNA was extracted using the RNAqueous Kit (Ambion) and Turbo DNase treatment (Ambion), according to the manufacturer's recommendations. RNA quality was verified with a Model 2100 Bioanalyzer (Agilent), and RNA concentration was determined using a RediPlate 96 RiboGreen RNA Quantitation Kit (Gibco, Invitrogen).
Gene expression was analysed using RT2 Profiler PCR Array Data Analysis software (version 3.4; SABiosciences). Changes in expression or activity levels are expressed as fold changes. Results represent the fold increase relative to the control samples (untreated DC).
Statistical analysis
All data are expressed as means with their standard errors. Statistical analyses were performed using the Mann–Whitney U test. Statistical calculations were made using NCSS 2007 software. P< 0·05 was considered as statistically significant.
Results
Lactobacillus rhamnosus CNCM I-4036 and its supernatant reduce pro-inflammatory cytokine production in Escherichia coli-challenged human dendritic cells
The immunomodulatory effects of live L. rhamnosus CNCM I-4036 and its supernatant were determined in vitro using human DC. The addition of E. coli (E. coli CECT 742, E. coli CECT 515 or E. coli CECT 729) or lipopolysaccharides induced significant changes in the profile of cytokines in DC. The treatments with live L. rhamnosus CNCM I-4036 and its supernatant markedly increased the levels of pro-inflammatory cytokines such as IL-6, IL-8, IL-12p40 (Fig. 1) and TNF-α (Fig. 2), compared with the untreated DC (control). In DC, L. rhamnosus CNCM I-4036 and its CFS exerted differential effects on the induction of cytokines.
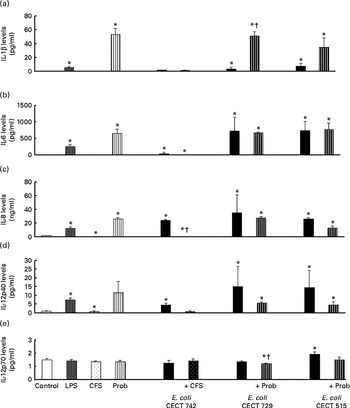
Fig. 1 Pro-inflammatory cytokine production in dendritic cells (DC) after exposure to Lactobacillus rhamnosus, Escherichia coli or both. DC incubated with different bacterial strains demonstrate distinct profiles of IL-1β (a), IL-6 (b), IL-8 (c), IL-12p40 (d) and IL-12p70 (e) production. The levels of these cytokines were measured after DC were incubated for 4 h with probiotics (live bacteria (Prob) or supernatant), pathogen or both and then further incubated for 20 h in media containing antibiotics. Culture supernatants were collected, and cytokine levels were assessed by immunoassay. Values are means of three different experiments, with their standard errors represented by vertical bars. * Mean value was significantly different from that of untreated DC (control) (P< 0·05). † Mean value was significantly different from that of pathogen-treated cells (P< 0·05). LPS, lipopolysaccharide; CFS, cell-free culture supernatant.
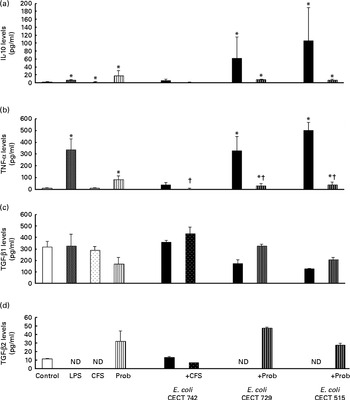
Fig. 2 Anti-inflammatory cytokine production in dendritic cells (DC) after exposure to Lactobacillus rhamnosus, Escherichia coli or both. DC incubated with different bacterial strains demonstrate distinct profiles of IL-10 (a), TNF-α (b), transforming growth factor (TGF)-β1 (c) and TGF-β2 (d) production. The levels of these cytokines were measured after DC were incubated for 4 h with probiotics (live bacteria (Prob) or supernatant), pathogen or both and then further incubated for 20 h in media containing antibiotics. Culture supernatants were collected, and cytokine levels were assessed by immunoassay. Values are means of three different experiments, with their standard errors represented by vertical bars. * Mean value was significantly different from that of untreated DC (control) (P< 0·05). † Mean value was significantly different from that of pathogen-treated cells (P< 0·05). LPS, lipopolysaccharide; CFS, cell-free culture supernatant; ND, not determined.
Interestingly, in the absence of any pathogen, the CFS by itself did not affect the secretion of pro-inflammatory cytokines or chemokines. Moreover, CFS treatment decreased the secretion of IL-8, IL-12p40 (Fig. 1) and all the chemokines tested compared with the untreated DC (control) (Fig. 3). In response to the stimulation with L. rhamnosus CNCM I-4036, supernatant and E. coli CECT 742, there was a decrease in the release of pro-inflammatory cytokines such as IL-8 and TNF-α (Figs. 1 and 2) and chemokines such as monocyte chemotactic protein-1 and RANTES (Fig. 3). Furthermore, in the presence of E. coli CECT 742, CFS induced the secretion of TGF-β1 (Fig. 2).
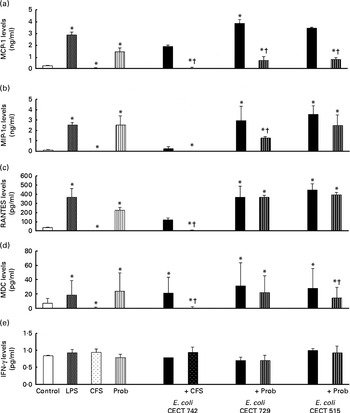
Fig. 3 Release of chemokines in dendritic cells (DC) after exposure to Lactobacillus rhamnosus, Escherichia coli or both. DC incubated with different bacterial strains demonstrate distinct profiles of monocyte chemotactic protein-1 (MCP-1)/chemokine (C–C motif) ligand (CCL)-2 (a), macrophage inflammatory protein-1α (MIP-1α)/CCL-3 (b), regulated upon activation, normal T-cell expressed and presumably secreted (RANTES)/CCL-5 (c), macrophage-derived chemokine (MDC)/CCL-22 (d) and interferon (IFN)-γ (e) production. The levels of these chemokines were measured after DC were incubated for 4 h with probiotics (live bacteria (Prob) or supernatant), pathogens or both and then further incubated for 20 h in media containing antibiotics. Culture supernatants were collected, and cytokine levels were assessed by immunoassay. Values are means of three different experiments, with their standard errors represented by vertical bars. * Mean value was significantly different from that of untreated DC (control) (P< 0·05). † Mean value was significantly different from that of pathogen-treated cells (P< 0·05). LPS, lipopolysaccharide; CFS, cell-free culture supernatant.
In general, as shown in Figs. 1 and 2, co-treatment with live L. rhamnosus CNCM I-4036 and E. coli had little effect on cytokine secretion, with the exception of a significant decrease in the levels of TNF-α and monocyte chemotactic protein-1 (Fig. 3). In contrast, in the absence of pathogens, live L. rhamnosus CNCM I-4036 increased the overall production of cytokines and chemokines, including that of IL-1β and IL-6 (Figs. 1–3). In the culture media, the analyses of TGF-β clearly showed that live L. rhamnosus CNCM I-4036 potently induced the production of TGF-β1 and TGF-β2 (Fig. 2). We did not detect TGF-β3 under any of the treatment conditions (data not shown).
Lactobacillus rhamnosus CNCM I-4036 induces Toll-like receptor signalling pathway gene expression in human dendritic cells
We also evaluated TLR signalling pathway gene expression. As shown in Figs. 4–7, exposure of DC to the different strains of E. coli for 4 h resulted in similar TLR expression patterns. The E. coli strains upregulated the gene expression of TLR-1, TLR-2 and TLR-4 (Fig. 4). Notably, TLR-9 was overexpressed in the treated DC (Fig. 5), and increased the gene expression of IL-1 receptor-associated kinase-4 (IRAK-4), c-Jun amino-terminal kinase (JNK), interferon regulatory factor (IRF-3) and IL-10 (Figs. 5–7).
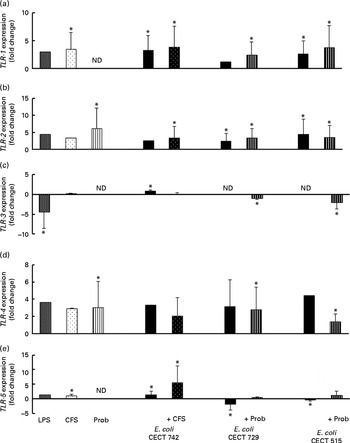
Fig. 4 Comparison of Toll-like receptor-1 (TLR-1) (a), TLR-2 (b), TLR-3 (c), TLR-4 (d) and TLR-5 (e) expression in dendritic cells (DC) in the presence of probiotic (Prob)/supernatant, Escherichia coli or both. The fold change represents the ratio of the expression in the treated DC compared with the control cells. Values are means of three different experiments, with their standard errors represented by vertical bars. * Mean value was significantly different from that of untreated DC (control) (P< 0·05). ND, not determined; LPS, lipopolysaccharide; CFS, cell-free culture supernatant.
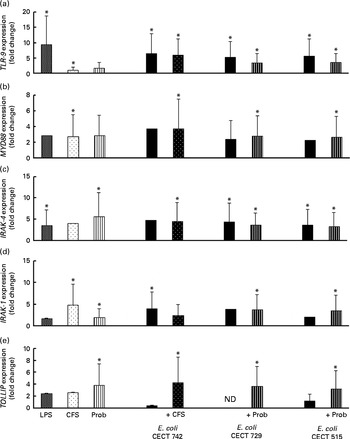
Fig. 5 Comparison of Toll-like receptor 9 (TLR-9) (a), myeloid differentiation factor 88 (MYD88) (b), IL-1 receptor-associated kinase 4 (IRAK-4) (c), IRAK-1 (d) and Toll-interacting protein (TOLLIP) (e) expression in dendritic cells (DC) in the presence of probiotic (Prob)/supernatant, Escherichia coli or both. The fold change represents the ratio of the expression in the treated DC relative to the control cells. Values are means of three different experiments, with their standard errors represented by vertical bars. * Mean value was significantly different from that of untreated DC (control) (P< 0·05). LPS, lipopolysaccharide; CFS, cell-free culture supernatant; ND, not determined.
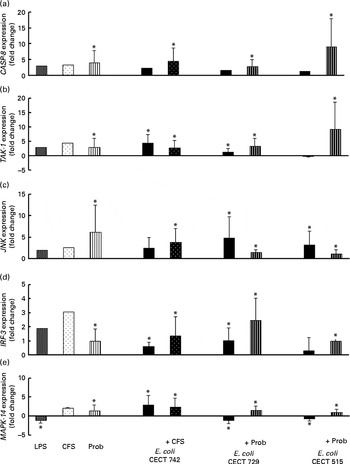
Fig. 6 Comparison of caspase-8 (CASP-8) (a), TGF-β-activated kinase-1 (TAK-1) (b), c-Jun amino-terminal kinase (JNK) (c), interferon regulatory factor-3 (IRF-3) (d) and mitogen-activated protein kinase-14 (MAPK-14) (e) expression in dendritic cells (DC) in the presence of probiotic (Prob)/supernatant, Escherichia coli or both. The fold change represents the ratio of the expression in the treated DC compared with the control cells. Values are means of three different experiments, with their standard errors represented by vertical bars. * Mean value was significantly different from that of untreated DC (control) (P< 0·05). LPS, lipopolysaccharide; CFS, cell-free culture supernatant.
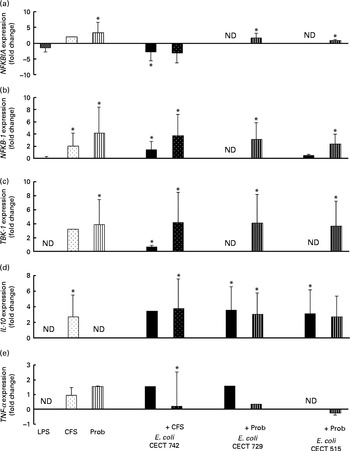
Fig. 7 Comparison of NF-κB inhibitor α (NFKBIA) (a), NFKB-1 (b), TANK-binding kinase-1 (TBK-1) (c), IL-10 (d) and TNF-α (e) expression in dendritic cells (DC) in the presence of probiotic (Prob)/supernatant, Escherichia coli or both. The fold change represents the ratio of the expression in the treated DC compared with the control cells. Values are means of three different experiments, with their standard errors represented by vertical bars. * Mean value was significantly different from that of untreated DC (control) (P< 0·05). ND, not determined; LPS, lipopolysaccharide; CFS, cell-free culture supernatant.
The probiotic strain L. rhamnosus CNCM I-4036 and its CFS similarly affected TLR pathway gene expression (Figs. 4–7). The probiotic strain alone (live L. rhamnosus) stimulated the gene expression of TLR-2 and TLR-4, whereas its supernatant significantly increased the expression of TLR-1 and TLR-5 (Fig. 4). Both treatments induced strong and sustained transcription of TLR-9 (Fig. 5).
Interestingly, the gene expression of TLR-1, TLR-2, TLR-5 and TLR-9 was notably increased in response to the stimulation with CFS and E. coli (Figs. 4 and 5). Conversely, DC exposure to live probiotic and enteropathogens down-regulated the gene expression of TLR-3, but increased the expression of TLR-4 (Fig. 4). Interestingly, the gene expression of TLR-5 was increased in response to the stimulation with CFS and E. coli (Fig. 4).
As shown in Figs. 4–7, exposure of DC to the probiotic strain L. rhamnosus CNCM I-4036 or its CFS with E. coli activated the TLR signalling pathway, and the expression of all the genes tested was increased.
Interestingly, upon stimulation with CFS or live probiotic plus E. coli, the gene expression of Toll-interacting protein (TOLLIP; Fig. 5) and caspase-8 (CASP-8; Fig. 6) was increased. A similar effect was observed for IRF-3 (Fig. 6) and TANK-binding kinase-1 (TBK-1; Fig. 7).
We also observed differences between the treatments. In the presence of E. coli, the CFS induced the expression of JNK, whereas its expression was decreased by L. rhamnosus (Fig. 6). In addition, the stimulation with CFS and E. coli decreased the gene expression of NF-κB inhibitor α (NFKBIA), whereas it was increased by the live probiotic strain (Fig. 7).
Discussion
Several studies have suggested that the in vitro peripheral blood mononuclear cell model is a useful screening tool to identify characteristic traits of probiotic strains to select probiotic strains for clinical trials( Reference Helwig, Lammers and Rizzello 17 – Reference Pérez-Cano, Dong and Yaqoob 21 ). However, these peripheral blood mononuclear cells and murine DC are quite different from human gut DC. Therefore, we employed an in vitro culture system using human DC that were harvested from umbilical cord blood CD34+ progenitor cells (and were similar to lamina propria gut DC) to study the immunological effects and anti-inflammatory properties of L. rhamnosus CNCM I-4036, a potentially probiotic strain isolated from the faeces of breast-fed newborn infants, and its supernatant.
Differences in the induction of the cytokine profile in human DC challenged with E. coli were observed between L. rhamnosus CNCM I-4036 and its supernatant. Specifically, in the presence of E. coli, L. rhamnosus CNCM I-4036 and its CFS reduced the production of pro-inflammatory cytokines and chemokines. However, the CFS did not induce the secretion of IL-12p40. In addition, in the absence of enteropathogens, the stimulation with CFS did not remarkably alter DC-produced cytokines. Recently, our group demonstrated a similar effect for L. paracasei CNCM I-4034( Reference Bermudez-Brito, Muñoz-Quezada and Gomez-Llorente 8 ) and Bifidobacterium breve CNCM I-4035 supernatants( Reference Bermudez-Brito, Muñoz-Quezada and Gomez-Llorente 22 ); the CFS from these probiotic strains exhibited anti-inflammatory behaviour by decreasing the production of inflammatory cytokines. Taken together, these data suggest that the probiotic strains L. paracasei CNCM I-4034, B. breve CNCM I-4035 and L. rhamnosus CNCM I-4036 release unknown factors capable of modulating DC cytokine production. One possible mechanism that could explain these immunomodulatory properties is the production and secretion of inhibitory metabolites in the media by probiotics. As a reflection of the differences between live probiotic bacteria and their supernatants, it is important to take into account that the supernatants were concentrated three times, so any substance released in the media would have a 10-fold effect compared with the live probiotic. In addition, these compounds that are present in different concentrations in live L. rhamnosus and its supernatant, but with the same origin, could explain the similar effects on TLR signalling genes. In contrast, the identification of proteins predicted to be involved in such interactions can pave the way towards well-targeted studies of the protein-mediated contacts between bacteria and the host, with the goal to enhance the understanding of the mode of action of probiotic bacteria. Ongoing work in our laboratory to unravel the proteins involved in these interactions reveals that these bacteria secrete a number of proteins that are predicted to play an important role in immunomodulation of the host immune system. Of special interest is the identification of three proteins excreted by L. rhamnosus CNCM I-4036 that may have potential implications in the direct interaction with the host, i.e. attachment to mucin and intestinal cells (M Fernández, S Juárez, S Ciordia, JM Vieites, JP Albar, A Gil and M Ferrer, unpublished results).
Interestingly, in the presence of E. coli, L. rhamnosus CNCM I-4036 promoted the production of TGF-β1 and TGF-β2, whereas the supernatant increased the secretion of TGF-β1. This effect was also observed with L. paracasei CNCM I-4034 and its supernatant( Reference Bermudez-Brito, Muñoz-Quezada and Gomez-Llorente 8 ). TGF-β1 is an important inhibitor of the synthesis of pro-inflammatory cytokines( Reference Parlesak, Haller and Brinz 23 ). This result indicates that the secretion of TGF-β1 after DC probiotic stimulation may be a mechanism by which these Lactobacillus strains exert anti-inflammatory effects.
The probiotic strain L. rhamnosus CNCM I-4036 did not reduce the secretion of IL-6, and, in fact, IL-6 induction by the probiotic strain was observed in the absence of any pathogen. IL-6 promotes terminal differentiation of B cells into plasma cells, and furthermore, it has been found to be able to polarise naive CD4+ T cells to effector Th2 cells. The relatively strong ability to induce the production of IL-6 that was observed for all the Lactobacillus strains is consistent with the general finding that many species of lactic acid bacteria induce IL-6 production and enhance intestinal IgA responses( Reference Christensen, Frøkiaer and Pestka 9 ).
In the absence of E. coli, the probiotic strain induced the production of IL-1β, IL-6, IL-8, IL-12 and TNF-α, whereas the supernatant suppressed the release of these pro-inflammatory cytokines. These results are in accordance with the earlier findings demonstrating that only intact bacteria induce appreciable amounts of IL-12( Reference Zeuthen, Fink and Frøkiaer 24 ). Because expression of IL-8, TNF-α and IL-6 is regulated by NF-κB( Reference Vossenkämper, Marchès and Fairclough 25 ), these results suggest that live L. rhamnosus CNCM I-4036 may enhance NF-κB-mediated innate immunity rather than suppression, as reported by Pagnini et al. ( Reference Pagnini, Saeed and Bamias 26 ) . Furthermore, NF-κB reportedly plays a beneficial role in epithelial cells. The activation of NF-κB similarly protects against the development of systemic inflammation( Reference Pagnini, Saeed and Bamias 26 ).
The probiotic L. rhamnosus CNCM I-4036 and its supernatant stimulated the gene expression of TLR-9 in the presence or absence of E. coli, indicating that this bacterial strain contains immunostimulatory double-stranded DNA motifs with unmethylated CpG sequences( Reference Plantinga, van Maren and van Bergenhenegouwen 27 ), which are powerful activators of the innate immune system. There are three types of CpG oligodeoxynucleotides, namely CpG-A, CpG-B and CpG-C (a mixture of CpG-A and CpG-B). CpG oligodeoxynucleotide strongly activates conventional DC to produce pro-inflammatory IL-6 and IL-12( Reference Ma, Muranyi and Chu 28 ). It is known that CpG oligodeoxynucleotide activates its receptor TLR-9, which in turn recruits the adaptor protein myeloid differentiation factor 88 and IRAK4, leading to the phosphorylation and activation of IRAK4( Reference Kumar, Kawai and Akira 29 ). In addition, it has been reported that Lactobacillus-mediated cytokine induction is strongly dependent on TLR-9( Reference Plantinga, van Maren and van Bergenhenegouwen 27 ).
As expected, DC expressed TLR-2 in response to the stimulation with L. rhamnosus. TLR-2 has been shown to recognise peptidoglycan, which is the main component of Gram-positive bacteria such as Lactobacillus. In addition, lactobacilli have been shown to mediate their effects via TLR-2( Reference Zeuthen, Fink and Frøkiaer 24 , Reference Maldonado Galdeano and Perdigón 30 ), possibly via lipoteichoic acid, a TLR-2 agonist and one of the main immunostimulatory components of Lactobacillus strains. Furthermore, in vitro and ex vivo studies have demonstrated that some immunostimulatory lactobacilli interact with intestinal epithelial cells and induce the release of IL-6 through an interaction with TLR-2( Reference Maldonado Galdeano and Perdigón 30 ). Interestingly, in the presence of E. coli and the supernatant, DC up-regulated the expression of TLR-5, and a previous study suggested that expression of this TLR may prevent immune responses against commensal Gram-negative bacteria( Reference Uematsu, Fujimoto and Jang 31 ).
Taken together, the present results demonstrate that exposure of intestinal-like human DC to the probiotic strain L. rhamnosus CNCM I-4036 or its CFS with E. coli activates the TLR signalling pathway, as the expression of each gene tested was increased, with the exception of TNF-α and NFKBIA. In the absence of pathogens, the probiotic strain L. rhamnosus CNCM I-4036 and its CFS increased the expression of NFKBIA, which is one of the most important negative regulators of NF-κB and acts by sequestering this transcription factor in the cytoplasm and preventing its nuclear translocation. These data reveal the anti-inflammatory character of L. rhamnosus CNCM I-4036 and its CFS. In addition, it has been reported that L. salivarius UCC118 induces the expression of this gene in Caco-2 cells( Reference O'Callaghan, Buttó and MacSharry 32 ).
In conclusion, the present results indicate that some probiotic strains such as L. rhamnosus CNCM I-4036 are immunostimulatory, whereas other strains are immunomodulatory( Reference Bermudez-Brito, Muñoz-Quezada and Gomez-Llorente 8 ), which demonstrates that generalisations concerning the biological effects and potential health benefits of probiotics should be avoided( Reference Williams 33 ) and implies that specific probiotic strains interact uniquely with DC to induce strain-specific effects( Reference Lebeer, Vanderleyden and De Keersmaecker 34 ).
Finally, L. rhamnosus CNCM I-4036-secreted products sensed by host pattern-recognition receptors exhibited an extraordinary ability to suppress the production of pro-inflammatory cytokines by DC and may be used as an effective and safer alternative to live bacteria.
Acknowledgements
The present study was supported by Hero Group, Spain S.A. through a contract (no. 3143) signed with the Fundación General Universidad de Granada-Empresa and co-sponsored by a CDTI project, a public entity of the Ministry of Economy and Competitiveness of Spanish Government. C. G.-L. has a postdoctoral fellowship from the Plan Propio of the University of Granada. Hero Group, Spain S.A. had no role in the design, analysis or writing of this article.
The authors' contributions are as follows: A. G. and C. G.-L. conceived and designed the experiments; M. B.-B. and S. M. Q. performed the experiments; M. B.-B. analysed the data; F. R. contributed reagents/materials/analysis tools; M. B.-B. and A. G. wrote the manuscript.
F. R. is a member of Hero Global Technology Centre for Infant Nutrition, Hero Group, Spain S.A. The sponsor had no role in the biological sample analysis, statistical analysis or data interpretation. None of the other authors has any conflict of interest.









