Obesity is one of the major underlying causes of the metabolic syndrome, including insulin resistance, type 2 diabetes and dyslipidaemia. Although the exact mechanisms by which obesity induces or worsens the metabolic risk factors are still unknown, excess adiposity, particularly visceral fat accumulation, is associated with insulin resistance and abnormal glucose and lipid metabolisms( Reference Jung and Choi 1 ). In addition, chronic low-grade inflammation in the adipose tissue contributes to the pathogenesis of insulin resistance and metabolic syndrome( Reference Jung and Choi 1 ).
Diet composition plays an important role in the development of obesity and its associated metabolic diseases( Reference Rolls and Hammer 2 ). Dietary fat is the most energy-dense macronutrient and causes less satiety than carbohydrate or protein( Reference Rolls and Hammer 2 ). Prolonged ingestion of high-fat diet (HFD) has been found to induce hyperphagia, body-weight gain and fat deposition and increase the levels of circulating glucose, insulin and TAG in rats( Reference Savastano and Covasa 3 – Reference Leibowitz, Dourmashkin and Chang 5 ). However, results have been inconsistent, with some studies reporting that HFD did not cause hyperphagia, hypertriglycerolaemia, hyperglycaemia or hyperinsulinaemia in mice( Reference Do, Oh and Kwon 6 , Reference Kalupahana, Voy and Saxton 7 ).
Carbohydrates, especially simple sugars, also seem to be associated with obesity and metabolic syndrome. Long-term ingestion of a sugar-rich diet increases fat mass without a concomitant increase in energy intake( Reference Oscai, Miller and Arnall 8 ) and promotes insulin resistance in rats( Reference Hulman and Falkner 9 ). Furthermore, a low-fat, high-carbohydrate diet had deleterious effects on cardiovascular health in obese adults compared with a high-fat, low-carbohydrate diet( Reference Santos, Esteves and da Costa Pereira 10 ). In contrast, another study has shown that a high-fat, low-carbohydrate diet decreases serum TAG levels and increases serum HDL-cholesterol levels in obese subjects, without concomitant changes in body weight or composition, in comparison with a low-fat, high-carbohydrate diet( Reference Ruth, Port and Shah 11 ). These results show that the role of diet in obesity varies with diet composition, animal species and experimental protocol.
Recently, nutritional genomics studies have analysed the responses of tissues to different diets and nutrients in order to provide a insight into the molecular events underlying diet-induced obesity( Reference Hageman, Wagener and Hantschel 12 ). However, obesity-related metabolic and molecular changes in response to HFD (containing low carbohydrate) v. low-fat diet (LFD; containing high carbohydrate) are not yet fully understood. Moreover, the HFD used in most animal studies of diet-induced obesity contains an extremely high fat content (approximately 60 % of total energy)( Reference Ruth, Port and Shah 11 ), which does not mimic the moderate fat content of Western human diets (approximately 40–45 % of total energy).
Therefore, the primary aim of the present study was to compare the effects of long-term ingestion of HFD (189 % of energy from fat) and LFD (which provides more carbohydrate, namely maize starch and sugar) on food intake, energy expenditure, body weight and adiposity, as well as plasma glucose, insulin, lipid and adipocytokine profiles, in C57BL/6J mice. The secondary aim of the present study was to obtain a further insight into the molecular mechanisms underlying the development of diet-induced obesity and its related metabolic abnormalities in response to HFD and to identify differentially expressed genes in the epididymal white adipose tissue (WAT) of HFD-fed mice using microarray analysis.
Experimental methods
Feeding protocol, energy expenditure and blood biomarkers
Male C57BL/6J mice (4-week old) were purchased from Jackson Laboratories and individually housed at room temperature on a 12 h light–12 h dark cycle. After 1 week of acclimation, they were fed a LFD (D12450B, 16·17 kJ/g; Research Diets) or a HFD (D12451, 19·866 kJ/g; Research Diets) ad libitum for 16 weeks. The LFD contains 42 % of energy from fat (25·2 % of energy from soyabean oil and 16·8 % of energy from lard), 294 % of energy from carbohydrate (130·2 % of energy from maize starch, 16·8 % of energy from maltodextrin and 147 % of energy from sucrose) and 84 % of energy from protein, whereas the HFD contains 189 % of energy from fat (25·2 % of energy from soyabean oil and 163·8 % of energy from lard), 147 % of energy from carbohydrate (29·4 % of energy from maize starch, 42 % of energy from maltodextrin and 75·6 % of energy from sucrose) and 84 % of energy from protein. Food intake of each mouse was measured daily throughout the study by subtracting the remaining food from the amount of food given to the mice, and daily food intake was calculated from the averaged food intake throughout the study. Energy expenditure for 24 h was measured at 14 weeks of feeding on the experimental diets using an indirect calorimeter (Oxylet; Panlab). Mice were killed as described previously( Reference Do, Oh and Kwon 6 ), and epididymal WAT, perirenal WAT, retroperitoneal WAT, mesenteric WAT and subcutaneous WAT were promptly removed, rinsed with physiological saline and weighed after blood collection. Among them, the epididymal WAT, which is widely used for the metabolic study due to its anatomically distinct feature, relative abundance and metabolic sensitivity( Reference Bjørndal, Burri and Staalesen 13 – Reference Liu, Shen and Ueno 18 ), was immediately fixed in 10 % buffered formalin for morphological examination and was frozen in liquid N2 and stored at − 70°C until RNA analysis. Plasma concentrations of NEFA, phospholipid (Wako Chemicals), TAG, total cholesterol and HDL-cholesterol (Asan Pharmaceutical Co., Ltd) were determined using commercially available kits.
Plasma adipocytokine and insulin levels were determined using a multiplex detection kit (Bio-Rad) and analysed using a Luminex 200 Labmap system. Fasting blood glucose concentration was measured using a glucose analyser (Glucocard; Arkray), and the homeostatic index of insulin resistance (HOMA-IR) was calculated as (fasting glucose (mmol/l) × fasting insulin (pmol/l)/135). All the experimental procedures were approved by the Kyungpook National University Ethics Committee (Approval no. KNU-2011-49).
Morphology of the epididymal white adipose tissue
The epididymal WAT was fixed in 10 % buffered formalin. Fixed tissues were embedded in paraffin, and 4 μm sections were prepared and stained with haematoxylin and eosin and Masson's trichrome. The stained areas were viewed using an optical microscope (Nikon) with a magnification of 200 × , and epididymal adipocyte size and fibrotic area from the Masson's trichrome staining were measured by computer analysis using the Leica Application Suite (version 2·8·1; Leica Microsystems).
RNA isolation, microarray analysis and real-time quantitative PCR
Total RNA was extracted from the epididymal WAT using TRIzol reagent (Invitrogen Life Technologies). For quality control, RNA purity and integrity were assessed using an Agilent 2100 Bioanalyzer (Agilent Technologies). Three pooled RNA sample sets were constructed to represent the LFD and HFD groups, as described previously( Reference Do, Kwon and Kim 19 ).
For microarray analysis, total RNA was amplified and purified using an Illumina RNA amplification kit (Ambion®) and quantified using an ND-1000 spectrophotometer (NanoDrop). Biotinylated cRNA (750 ng) was hybridised to MouseWG-6 v2·0 Expression BeadChips (Illumina, Inc.) at 58°C for 16–18 h. Array signal detection was carried out using Amersham Cy3-streptavidin (GE Healthcare Bio-Sciences). BeadChips were scanned using an Illumina BeadArray Reader, and raw data were extracted using the Illumina BeadStudio software. Probe signal intensities were quantile-normalised and log-transformed. Limma was used to determine significantly differentially expressed genes based on a false discovery rate less than 5 %, a Benjamin and Hochberg-adjusted P value < 0·05 and a log2 fold change greater than 1( Reference Do, Kwon and Kim 19 ). The DAVID Functional Annotation Tool was used to identify the enriched biological themes and cluster-redundant annotation terms. These microarray data were deposited in Gene Expression Omnibus database (accession no. GSE63198).
To validate microarray data, several differentially expressed genes (Rgs1, Mmp2, Ccl2, Tlr2, Tlr4 and Irs2) were measured independently by real-time quantitative PCR using the same pooled RNA samples that were hybridised to BeadChips. Total RNA (1 μg) was reverse-transcribed into complementary DNA using a QuantiTect reverse transcription kit (Qiagen), and real-time quantitative PCR was carried out on a CFX96 real-time system (Bio-Rad) using a SYBR Green PCR kit (Qiagen). Values were normalised to glyceraldehyde-3-phosphate dehydrogenase levels, and relative gene expression was calculated by the
![]() $$2^{ - \Delta \Delta Ct} $$
method.
$$2^{ - \Delta \Delta Ct} $$
method.
For definition of gene abbreviations, see online Supplementary Table S1.
Statistical analyses
All data are presented as means with their standard errors. Statistical analyses were performed using SPSS software. Changes in body weight were analysed by repeated-measures ANOVA, and other data were analysed by Student's t test or Wilcoxon t test. Results were considered statistically significant when P< 0·05.
Results
High-fat diet decreased food intake but increased body weight and fat mass by decreasing energy expenditure
HFD-fed mice exhibited a significantly greater body weight than LFD-fed mice, although the average daily food intake throughout the study was markedly lower in HFD-fed mice and the average daily energy intake did not differ between the two groups (Fig. 1(a)–(c)). Interestingly, indirect calorimetry revealed that HFD-fed mice had lower energy expenditure than LFD-fed mice (Fig. 1(d)). As expected, the subcutaneous WAT and the visceral WAT (including epididymal, perirenal, retroperitoneal and mesenteric WAT) weights, adipocyte size and adipose collagen accumulation were higher in HFD-fed mice (Fig. 1(e) and (f)).

Fig. 1 Metabolic and morphological phenotype of high-fat diet (HFD, ![]() ,
, ![]() )-fed mice. Values are means with their standard errors represented by vertical bars (n 10). (a) *** Mean value was significantly different from that of low-fat diet (LFD,
)-fed mice. Values are means with their standard errors represented by vertical bars (n 10). (a) *** Mean value was significantly different from that of low-fat diet (LFD, ![]() ) group (P< 0·001; repeated-measures ANOVA). (b–i) Mean value was significantly different from that of low-fat diet (LFD,
) group (P< 0·001; repeated-measures ANOVA). (b–i) Mean value was significantly different from that of low-fat diet (LFD, ![]() ) group: † P< 0·05, †† P< 0·01, ††† P< 0·001 (Student's t test). (f) Representative photographs of adipocytes in the epididymal white adipose tissue (WAT) of mice at × 200 magnification (left panel) and quantitative analysis (right panel). The WAT section stained with Masson's trichrome showed significant deposition of collagens, primarily collagen I and III (blue stain indicated with arrowheads), in HFD-fed mice. BW, body weight; H&E, haematoxylin and eosin; HOMA-IR, homeostatic index of insulin resistance. (A colour version of this figure can be found online at http://www.journals.cambridge.org/bjn)
) group: † P< 0·05, †† P< 0·01, ††† P< 0·001 (Student's t test). (f) Representative photographs of adipocytes in the epididymal white adipose tissue (WAT) of mice at × 200 magnification (left panel) and quantitative analysis (right panel). The WAT section stained with Masson's trichrome showed significant deposition of collagens, primarily collagen I and III (blue stain indicated with arrowheads), in HFD-fed mice. BW, body weight; H&E, haematoxylin and eosin; HOMA-IR, homeostatic index of insulin resistance. (A colour version of this figure can be found online at http://www.journals.cambridge.org/bjn)
High-fat diet induced hyperglycaemia, hyperinsulinaemia and insulin resistance
Fasting blood glucose level was significantly increased in HFD-fed mice (Fig. 1(g)). HFD also resulted in significant increases in plasma insulin level and HOMA-IR compared with LFD (Fig. 1(h) and (i)).
High-fat diet increased plasma leptin, resistin and plasminogen activator inhibitor type 1 levels but did not alter plasma adiponectin and cytokine levels
Plasma leptin, resistin and plasminogen activator inhibitor type 1 levels were significantly higher in HFD-fed mice than in LFD-fed mice (Table 1). However, there was no significant difference in plasma adiponectin level between the two groups. The levels of plasma cytokines (monocyte chemotactic protein 1, TNF-α, IL-6 and IL-10) were also not significantly altered by HFD.
Table 1 Plasma adipocytokine and lipid levels (Mean values with their standard errors; n 10)
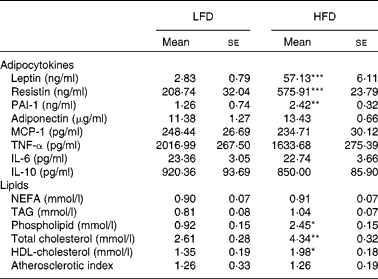
LFD, low-fat diet; HFD, high-fat diet; PAI-1, plasminogen activator inhibitor type 1; MCP, monocyte chemotactic protein.
Mean values were significantly different from those of the LFD group: * P< 0·05, ** P< 0·01, *** P< 0·001.
High-fat diet increased plasma phospholipid, total cholesterol and HDL-cholesterol levels but did not alter plasma NEFA and TAG levels
No significant differences in plasma NEFA or TAG levels were observed between the HFD and LFD groups (Table 1). However, plasma phospholipid level was significantly higher in HFD-fed mice. HFD also increased plasma total cholesterol level, as well as HDL-cholesterol level, resulting in no significant difference in atherogenic index between the two groups.
Gene expression profiles in the epididymal white adipose tissue
To determine the changes in global gene expression profiles of the epididymal WAT in HFD-induced obesity, we identified differentially expressed genes in HFD-fed mice compared with LFD-fed mice using microarray analysis. Of the 45 000 analysed expression probes, 1270 were different between the two groups. Among these 1270 HFD-responsive genes, 657 were up-regulated and 397 were down-regulated. The top ten differentially expressed genes are listed in Table 2. Functional annotation clustering using DAVID revealed that the majority of genes up-regulated by HFD were related to immune and inflammatory responses (Table 3; online Supplementary Table S2). These included genes encoding chemokines, receptors of chemokines and cytokines, toll-like receptors, C-type lectin receptors, Fc receptors and surface markers of immune cells (Table 4). Moreover, HFD induced up-regulation of genes encoding extracellular matrix (ECM) components such as collagen (Col1a1, Col3a1, Col5a2, Col6a2 and Col6a3), glycosaminoglycan and proteoglycan (Sdcbp and Lum), adhesive glycoproteins (Mfap5, Gpnmb and Fn1) and integrin (Itgad, Itgam and Itgax), as well as genes encoding proteins involved in ECM remodelling and regulation, such as cathepsins (Ctsa, Ctsh, Ctsk, Ctsl, Ctss and Ctsz), a disintegrin and metalloprotease (ADAM) domain (Adam8, Adam12 and Adam17), matrix metalloproteinases (MMP: Mmp2, Mmp3, Mmp12 and Mmp13), tissue inhibitors of metalloproteinases (Timp1) and other fibrosis-related genes (Tgfb1). Interestingly, the expression of two ADAM and MMP genes, Adam7 and Mmp9, was down-regulated in obesity.
Table 2 Top ten differentially expressed genes in the epididymal white adipose tissue of high-fat diet-fed mice
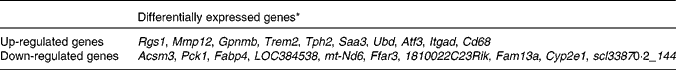
* Differentially expressed genes were determined using Limma in R/Bioconductor, based on P< 0·05, false discovery rate < 5 % and log2 fold change >1. For definition of gene abbreviations, see online Supplementary Table S1.
Table 3 Functional annotation clusters of up-regulated and down-regulated genes in the epididymal white adipose tissue of high-fat diet-fed C57BL/6J mice*

ES, enrichment score.
* Functional annotation terms were clustered according to biological processes.
Table 4 Fold changes of selected genes influenced by high-fat diet*

* Differentially expressed genes were determined using Limma in R/Bioconductor, based on P< 0·05, false discovery rate < 5 % and log2 fold change >1. For definition of gene abbreviations, see online Supplementary Table S1.
The genes that were down-regulated in response to HFD were enriched in gene ontology categories related to oxidation–reduction, fatty acid metabolism, insulin response and skeletal system development (Table 3; online Supplementary Table S2). Down-regulated genes related to oxidation–reduction included genes controlling antioxidant defence (Gpx3), detoxification (Aldh1a1, Aldh6a1, Cyp2d9, Cyp2d22, Cyp2e1, Cyp2f2, Gsta3 and Gsta4) and mitochondrial energy transduction pathways such as the TCA cycle (Por) and oxidative phosphorylation (Ndufb4 and Ndufb9) (Table 4). HFD decreased the expression of genes involved in lipolysis, thermogenesis (Adrb3) and fatty acid uptake, transport (Fabp4, Cd36 and Slc27a2), elongation (Elovl6), activation and oxidation (Acsm3, Acacb, Acot4, Acadsb, Hadh and Faah). Consistent with increased insulin resistance, insulin sensitivity-related genes, such as those encoding insulin receptor substrate 2 (Irs2) and GLUT 3 (Slc2a3), were down-regulated by HFD feeding. Along with Mmp9, multiple genes controlling skeletal system development and pattern specification (Bmp4, Hoxa5, Hoxa7, Hoxb5, Hoxc6, Irx3 and Igf2) were also down-regulated in HFD-fed mice. The expression of many genes encoding G-protein-coupled receptors (GPCR: Gpr64, Gpr120, Gpr109a, Gpr156 and Opn3) was decreased in response to HFD, whereas mRNA expression of regulators of G protein signalling (RGS: Rgs1 and Rgs10) was increased. Several genes identified as differentially expressed based on microarray analysis were independently validated by real-time quantitative PCR (Fig. 2).

Fig. 2 Validation of microarray data using real-time quantitative PCR (RT-qPCR) for (a) Rgs1, (b) Mmp2, (c) Ccl2, (d) Tlr2, (e) Tlr4 and (f) Irs2. Values are means with their standard errors represented by vertical bars. * P< 0·05, *** P< 0·001 (Wilcoxon t test). † P< 0·05, ††† P< 0·001 (Student's t test). LFD, low-fat diet; HFD, high-fat diet. For definition of gene abbreviations, see online Supplementary Table S1.
Discussion
It is generally accepted that HFD induces hyperphagia, and that increased energy intake is the major mechanism by which HFD causes obesity( Reference Rolls and Hammer 2 , Reference Savastano and Covasa 3 ). In the present study, however, despite lower food intake and similar energy intake, mice fed HFD (189 % of energy from fat) for 16 weeks gained more body weight and body fat than mice fed LFD (42 % of energy from fat), which provided more energy as carbohydrate (sugar and maize starch). This is consistent with a previous finding that body weight and adiposity, but not energy intake, increased in HFD-fed C57BL/6J mice compared with LFD-fed mice during the first 17 weeks of feeding, although energy intake was significantly higher in HFD-fed mice after 17 weeks owing to a reduction in leptin sensitivity( Reference Lin, Thomas and Storlien 20 ). Body weight is normally maintained by a balance between energy intake and energy expenditure. When energy intake exceeds energy expenditure, excess energy is stored as TAG in the adipose tissue, resulting in overweight or obesity. Because energy expenditure during dark hours was significantly lower in HFD-fed mice than in LFD-fed mice, the increase in body weight and fat mass in HFD-fed mice may be due to a positive energy balance resulting from a decrease in energy expenditure relative to energy intake.
Induction of energy expenditure based on fatty acid oxidation in WAT may reduce adiposity( Reference Flachs, Rossmeisl and Kuda 21 ). Mitochondrial biogenesis and thermogenesis are decreased in WAT of obese individuals and rodents( Reference Flachs, Rossmeisl and Kuda 21 – Reference Böttcher and Fürst 23 ), while induction of mitochondria and activation of fatty acid oxidation have been observed in WAT under conditions promoting loss of adiposity( Reference Flachs, Rossmeisl and Kuda 21 ). Kusminski and Scherer( Reference Kusminski and Scherer 24 ) suggested that in the obese state, mitochondria in WAT cannot cope with increasing demands for fatty acid oxidation, resulting in incomplete β-oxidation. In the present study, HFD down-regulated the expression of genes involved in fatty acid catabolism and oxidation, as well as genes controlling the mitochondrial energy transduction pathways, including the TCA cycle and oxidative phosphorylation, in the epididymal WAT. In other studies, expression of Faah (a catabolic gene for bioactive fatty acid amides) in adipose tissue was negatively correlated with visceral fat mass in human subjects( Reference Blüher, Engeli and Klöting 25 ), and mice lacking Faah had increased body weight and fat mass with decreased energy expenditure( Reference Vaitheesvaran, Yang and Hartil 26 ). Thus, down-regulation of genes regulating fatty acid catabolism, fatty acid oxidation and energy metabolism in epididymal WAT may be partly responsible for the decrease in energy expenditure induced by HFD. Our findings also suggest that the deleterious effects of HFD on adipose tissue mitochondria are associated with decreased expression of genes involved in antioxidant defence and detoxification. An altered redox state is associated with the accumulation of products of incomplete β-oxidation and increased mitochondrial reactive oxygen species formation, leading to a deterioration of insulin sensitivity( Reference Muoio and Neufer 27 ). Mitochondrial reactive oxygen species produced by redox imbalance may promote adipocyte differentiation( Reference Tormos, Anso and Hamanaka 28 ). In adipose tissue of obese mice such as KKAy mice and HFD-induced obese mice, reactive oxygen species production was accompanied by decreased expression of antioxidant enzymes( Reference Furukawa, Fujita and Shimabukuro 29 ).
In the present study, another candidate gene for controlling energy expenditure is the bone morphogenetic protein 4 (Bmp4) gene. Recently, Qian et al. ( Reference Qian, Tang and Li 30 ) reported that adipose-specific overexpression of Bmp4 led to reduced body fat mass and increased energy expenditure, whereas Bmp4-deficient mice exhibited increased adiposity and impaired insulin sensitivity, which suggests that Bmp4 can regulate WAT remodelling and induction of brown adipocyte-like cell structure and function. Interestingly, along with Bmp4, multiple genes involved in skeletal system development and pattern specification were down-regulated in the present study. Similarly, down-regulation of homeobox genes, including Hoxa5, was observed in the epididymal WAT of ob/ob mice compared with lean controls( Reference Yamamoto, Gesta and Lee 31 ), whereas expression of homeobox genes (e.g. Hoxa5, Hoxb5, Hoxc6 and Irx3) was up-regulated after fat loss in human subjects( Reference Dankel, Fadnes and Stavrum 32 ). Recently, Gehrke et al. ( Reference Gehrke, Brueckner and Schepky 33 )suggested epigenetic regulation, in particular DNA methylation of HOX genes as a mechanism that explains how these transcriptional factors exhibit such a distinct depot-specific expression pattern. Different adipose depots are known to mediate different effects on the risk for metabolic disorders( Reference Gehrke, Brueckner and Schepky 33 ). Moreover, obese patients with type 2 diabetes had lower expression of Igf2 in the adipose tissue compared with those who were lean( Reference Chen, Macpherson and Owens 34 ), and down-regulation of Igf2 in the adipose tissue contributed to the paternal transmission of HFD-induced obesity( Reference Morita, Horii and Kimura 35 ). However, many genes related to skeletal system development and pattern specification still have unknown roles in the development of obesity. Thus, understanding these genes will facilitate the development of novel approaches in the treatment of obesity.
In contrast, since the NEFA released by lipolysis or taken up by cells can be utilised in WAT as a source of energy through β-oxidation in the mitochondria, a decrease in the expression of genes involved in lipolysis and fatty acid uptake and transport in response to HFD may reduce β-oxidation, resulting in excessive fat accumulation. In mice, Adrb3 disruption reduces lipolysis and increases body fat accumulation( Reference Revelli, Preitner and Samec 36 ), and Adrb3 increases thermogenesis in the brown adipose tissue( Reference Arner and Hoffstedt 37 ). Deficiency of Fabp4 also reduces lipolysis in mice( Reference Scheja, Makowski and Uysal 38 ), and a decrease in Fabp4 expression in the adipose tissue has been found to be inversely associated with obesity in human subjects( Reference Queipo-Ortuño, Escoté and Ceperuelo-Mallafré 39 ).
The expression of several GPCR genes, including Gpr64, Gpr120, Gpr109a, Gpr156 and Opn3, was down-regulated by HFD in the present study. Mice lacking Gpr120, an NEFA-sensing GPCR, exhibited obesity, glucose intolerance and fatty liver( Reference Ichimura, Hirasawa and Poulain-Godefroy 40 ). Furthermore, Gpr109a has been reported to act as a metabolic sensor activated by intermediates of energy metabolism, and its expression was down-regulated in the adipose tissue of HFD-fed mice( Reference Wanders, Graff and Judd 41 ). In contrast, the expression of genes encoding RGS (Rgs1 and Rgs10) was up-regulated by HFD. The RGS family is involved in the rapid turn-off of GPCR signalling pathways( Reference De Vries, Zheng and Fischer 42 ). Huang et al. ( Reference Huang, Charbeneau and Fu 43 ) demonstrated that RGS-insensitive Gαi2G184S mice exhibited resistance to the metabolic effects of HFD, and were protected from insulin resistance by increasing energy expenditure and peripheral insulin sensitivity. Furthermore, in another study, Rgs2- and Rgs5-deficient mice had less body mass and body fat than littermate controls( Reference Cho, Park and Hwang 44 , Reference Nunn, Zhao and Zou 45 ). However, the specific roles of most GPCR and RGS genes in the adipose tissue are not yet fully understood.
Obesity is characterised by extensive reorganisation of the adipose tissue, which involves ECM remodelling and adipogenesis( Reference Crandall, Hausman and Kral 46 ). Cathepsins, ADAM, MMP and their inhibitors (i.e. TIMP) are involved in ECM remodelling( Reference Järveläinen, Sainio and Koulu 47 ). The present study indicated that mRNA levels of most genes involved in ECM remodelling were up-regulated in obese adipose tissues compared with lean tissues, while expression of Adam7 and Mmp9 was down-regulated in obesity. Interestingly, mice lacking Ctsk, Ctsl, Adam12, Timp1 or Mmp2, but not Mmp9, were protected against obesity( Reference Lijnen, Demeulemeester and Van Hoef 48 – Reference Van Hul, Piccard and Lijnen 53 ). Belo et al. ( Reference Belo, Souza-Costa and Luizon 54 ) reported the association between Mmp9 gene polymorphisms and lower plasma Mmp9 level in obese children and adolescents, suggesting a potential role of Mmp9 gene in the development of obesity. We also observed that HFD up-regulated the expression of Tgfb1 (encoding a profibrotic cytokine), as well as genes encoding various ECM components, including members of the collagen family, and increased adipose collagen accumulation, suggesting that HFD may facilitate abnormal ECM accumulation in WAT, promoting fibrosis. In addition, we detected significant expression of genes for a lymphocyte chemotactic factor (Cxcl12) and its receptor (Cxcr4), as well as genes related to macrophage infiltration (Ccl2, Cxcl14 and Ccr5) in WAT of HFD-fed mice. It is well known that macrophage infiltration plays a critical role in adipose tissue inflammation, and that chemokines are involved in this process. In obese mice, a deficiency of Ccl2, Cxcl14 or Ccr5 decreased macrophage infiltration in the adipose tissue and improved metabolic function, while overexpression of Ccl2 in the adipose tissue caused the opposite phenotype( Reference Kanda, Tateya and Tamori 55 – Reference Kitade, Sawamoto and Nagashimada 58 ). Besides macrophages, T and B lymphocytes have been shown to accumulate in the adipose tissue of obese mice and have been associated with inflammation and insulin resistance( Reference Kintscher, Hartge and Hess 59 , Reference Winer, Winer and Shen 60 ).
Many other genes involved in innate or adaptive immune and inflammatory responses, such as those encoding toll-like receptors, C-type lectin receptors, Fc receptors, cytokine receptors and triggering receptors, were up-regulated by HFD. One of these genes, Tlr2, has been associated with increased adiposity, macrophage infiltration and inflammatory cytokine expression( Reference Himes and Smith 61 ). Saa3, which encodes an inflammatory adipocytokine and may be a potential link between obesity and its metabolic complications( Reference Yang, Lee and Hu 62 ), was also one of the top ten genes up-regulated by HFD in the present study. However, despite the increased expression of multiple inflammatory genes in WAT of HFD-fed mice, plasma concentrations of inflammatory chemokines and cytokines were not affected, as described in a previous study( Reference Thomas, Dunn and Oort 63 ). Thomas et al. ( Reference Thomas, Dunn and Oort 63 ) suggested that shifts in these inflammation markers in the blood are probably not as sensitive as WAT mRNA patterns, which reflect the inflammatory status of the local tissue.
Tph2, Ubd and Atf3 were also among the genes most highly up-regulated by HFD in the present study. Although a deficiency of Tph2 or Ubd in mice caused body fat reduction by increasing energy expenditure( Reference Yadav, Oury and Suda 64 , Reference Canaan, Defuria and Perelman 65 ), the specific functions of these genes in the adipose tissue are still unclear. Atf3 has been reported to act as a negative regulator of SFA/TLR4 signalling and macrophage activation in obese adipose tissue( Reference Suganami, Yuan and Shimoda 66 ); however, overexpression of Atf3 decreased mitochondrial function and mitochondria-related gene expression in adipocytes in vivo and in vitro ( Reference Jang, Son and Jung 67 ). Thus, further experiments are required to evaluate whether Atf3 has a beneficial or detrimental role in obesity and its associated metabolic disturbances.
In conclusion, long-term HFD feeding predominantly up-regulated a series of genes involved in immune and inflammatory responses, as well as cell activation and division in WAT of C57BL/6J mice. Genes down-regulated by HFD were mainly implicated in fatty acid metabolism, oxidation–reduction, insulin response and skeletal system development. The top ten up-regulated genes were Rgs1, Mmp12, Gpnmb, Trem2, Tph2, Saa3, Ubd, Atf3, Itgad and Cd68, which might be associated with energy expenditure (Rgs1, Tph2, Ubd and Atf3), ECM remodelling and components (Mmp12, Gpnmb and Itgad) and inflammation (Trem2, Saa3, Atf3 and Cd68); further studies are needed to evaluate the roles of these genes in the pathophysiology of the adipose tissue. The functions of the top ten down-regulated genes, except Pck and Fabp4, are also poorly understood. Thus, the present findings will be useful in future efforts to identify the therapeutic targets for obesity. However, further studies are needed to investigate the molecular mechanisms responsible for WAT depot-specific differences.
Supplementary material
To view supplementary material for this article, please visit http://dx.doi.org/10.1017/S0007114515000100
Acknowledgements
The present study was supported by the Basic Science Research Program (U. J. J., grant no. NRF-2011-0022387, NRF-2014R1A1A4A01007858), the SRC program (M.-S. C., Center for Food & Nutritional Genomics, grant no. 2008-0062618) and the fundamental technology program (M.-S. C., grant no. 2012M3A9C4048818) of the National Research Foundation of Korea funded by the Ministry of Education, Science and Technology. This study received no specific grant from any funding agency, commercial or not-for-profit sectors. In addition, all the funders had no role in the design and analysis of the study or in the writing of this article.
The authors' contributions are as follows:
M.-S. C. designed the study and participated in the writing of the manuscript; Y.-J. K. performed the animal experiments; J. Y. R. and S. R K. were involved in data interpretation and edited the manuscript; U. J. J. analysed the data and wrote the manuscript. All authors read and approved the final manuscript.
The authors declare that there is no conflict of interest.









