Human milk is considered the optimal form of nourishment for infants during the first 6 months of life( 1 ). Human milk is a complex biological fluid containing a wide variety of nutrients and bioactive molecules that contribute to a healthy infant growth and development( Reference Hill and Newburg 2 ). The composition of breast milk is not constant; some factors known to influence breast milk composition include stage of lactation, parity, gestational age, maternal diet, time of day and time into feeding( Reference Ballard and Morrow 3 – Reference Sinanoglou, Cavouras and Boutsikou 5 ). Although some of these variations are physiological and thought to adapt to the needs of the infant (e.g. variations during lactation), other variations (e.g. those resulting from a non-optimal maternal diet) may affect milk’s nutritional value and cause child malnutrition, including undernutrition or overnutrition( Reference Shahrin, Chisti and Ahmed 6 ).
Nutrients should not be considered only as a source of energy or as factors involved in the development of the organism; nutrients are also able to interact with specific genes( Reference Verduci, Banderali and Barberi 7 ). Breast milk fatty acids (FA), for instance, provide energy to the breast-fed infant but can also act as structural elements of cell membranes, be precursors of inflammatory mediators and influence gene expression( Reference Richard, Lewis and Field 8 – Reference Mennitti, Oliveira and Morais 10 ). In recent years, a new field of study has developed, which combines two different areas of research called nutrigenomics and nutrigenetics. Nutrigenomics explains how nutrients are, either directly or indirectly, able to influence gene expression. Nutrigenetics explains how genes are able to modulate the individual response to nutrients( Reference Fenech, El-Sohemy and Cahill 11 ). Through both areas of research it may be possible to identify mechanisms that underlie individual variations in dietary requirements, as well as in the capacity to respond to food-based interventions( Reference Verduci, Banderali and Barberi 7 ).
In this context, the present review is conducted to understand how maternal FA intake modulates the composition of milk fat by interacting with maternal genes (nutrigenomics), and how maternal genes modulate the effect of maternal FA intake on milk fat (nutrigenetics). First, we describe the role and origin of breast milk FA, and then we provide information on the importance of nutrigenomics and nutrigenetics in influencing breast milk’s FA composition.
Methods
Information on the mammary gland development, the role and sources of human milk FA and the role of genetic polymorphisms on milk from ruminant animals was obtained from observational and intervention studies and (systematic) reviews gathered via the electronic databases PubMed platform, EMBASE, Cochrane Library and Medline.
For the sections of nutrigenomics and nutrigenetics of breast milk FA, we undertook a comprehensive review of the literature via the same above-mentioned databases for the period 2005–2017 using the terms ‘nutrigenetics’, ‘nutrigenomics’, ‘human milk composition’, ‘fatty acids’, ‘breast-feeding’, ‘maternal diet’, ‘polymorphisms’, ‘gene-diet interaction’ and ‘gene expression’. Some older references were also included if they were needed to provide background knowledge (e.g. Nara et al.( Reference Takeuchi, Yahagi and Izumida 52 ) were among the first researchers to identify the sterol-regulatory elements (SRE) in the human fatty acid desaturase (FADS2) promoter). Moreover, a study carried out by Del Prado et al.( Reference Liu, Liu and Wu 31 ) provided an overview of the various factors (diet, endogenous synthesis and body reserves mother) that may influence the composition of breast milk. The fundamental inclusion criterion was experimental and updated studies published in English.
Mammary gland development and human milk fatty acids
Mammary gland development
Human milk is produced in the mammary gland, which is composed of glandular tissue (lactocytes), connective and adipose tissue and vascular stroma. The mammary epithelium is composed primarily of two types of differentiated epithelial cells: luminal secretory cells and a layer of basal myoepithelial cells. A network of branching longitudinal striated cells called myoepithelial cells surrounds the alveoli and the smaller ducts( Reference Arendt and Kuperwasser 12 ). Contraction of these cells squeezes the alveoli and causes ejections of milk into the main duct (lactiferous) localised behind the nipple( Reference Gardner, Kent and Lai 13 ).
Mammary gland development begins at puberty and continues throughout pregnancy until lactation. There are four defined stages involved in the mammary gland development: mammogenesis, lactogenesis, galactopoiesis and involution( Reference Riordan 14 ). The proportion of macronutrients and micronutrients from human milk progressively changes with the onset and duration of lactation. There are three phases of milk: colostrum, transitional and mature milk, each with distinct characteristics( Reference Ballard and Morrow 3 , Reference Golinelli, Del Aguila and Flosi Paschoalin 15 ).
Role of milk fatty acids
Among the major components of human milk, fat is the second largest component and has a very important role in infant’s development as a source of energy and essential lipids( Reference Innis 16 , Reference Koletzko, Agostoni and Bergmann 17 ). The main components of milk fat are FA, which are mainly found in the form of TAG. TAG account for 98 % of milk fat( Reference Mohammad and Haymond 18 ). FA can be saturated or unsaturated depending on the presence of double bonds. The importance of fat in breast milk as a main source of energy and essential lipid-soluble vitamins and in optimum infant development has been reported( Reference Ballard and Morrow 3 , Reference Andreas, Kampmann and Mehring Le-Doare 9 , Reference Koletzko 19 ). FA also participate in transcriptional regulation of biosynthesis of lipoproteins from the liver( Reference Vallim and Salter 20 ), promoting successful lipoprotein metabolism in infants. On the other hand, n-3 long-chain PUFA (n-3 LC-PUFA) are important components of the brain and the retina( Reference Delgado-Noguera, Calvache and Bonfill Cosp 21 − Reference Ryan, Astwood and Gautier 23 ) and their deficiency during brain maturation reduces plasticity and compromises brain function in adulthood( Reference Bhatia, Agrawal and Sharma 24 ). Several studies suggest that maternal intake of omega-3 LC-PUFA during pregnancy may have favourable effects enhancing cognitive functions and attention in the infant( Reference Smithers, Gibson and McPhee 25 , Reference Jacobson, Jacobson and Muckle 26 ).
Sources of breast milk fatty acids
The FA secreted in breast milk have different origins: FA up to C14 : 0 originate from de novo synthesis in the breast, whereas FA with a chain length greater than C14 : 0 originate from the maternal diet or body stores( Reference Nasser, Stephen and Goh 27 ). Linoleic acid (LA) and α-linolenic acid (ALA) are essential FA, which means that they cannot be synthesised in the human body and therefore need to be obtained through the diet. The rest of the FA can be synthesised de novo and are therefore considered non-essential. MUFA contain a single double bond( Reference Schwingshackl and Hoffmann 28 ) and LC-PUFA have more than eighteen to twenty carbon atoms and more than one double bond; they can be synthesised de novo, mobilised from adipose tissue or obtained from dietary fat (e.g. olive oil, meat, fish)( Reference Schwingshackl and Hoffmann 28 − Reference Rudolph, Monks and Burns 30 ). The role of the de novo synthesis, diet and mobilisation of body stores are described shortly in the following paragraphs.
De novo synthesis
De novo synthesis of SFA and MUFA involves the enzyme fatty acid synthase (FAS) and stearoyl-CoA desaturase (SCD), respectively( Reference Liu, Liu and Wu 31 , Reference Green, Ozguden-Akkoc and Wang 32 ). PUFA can be classified into two main families, n-6 and n-3, depending on the position of the first double bond counting from the methyl end group of the FA( Reference Abedi and Sahari 33 ). As mentioned above, LA and ALA cannot be synthesised de novo by humans and therefore need to be supplied through the diet. The remaining n-6 and n-3 PUFA can be synthesised from the precursors LA and ALA through a series of elongation and desaturation reactions. LA is metabolised to AA, whereas ALA is metabolised to EPA and DHA as active metabolic products( Reference Bazinet and Layé 34 ) (Fig. 1).

Fig. 1 Synthesis of long-chain PUFA from linoleic acid or α-linolenic acid, showing the steps catalysed by Δ5 and Δ6 desaturases. ![]() , Routes not known to occur in humans.
, Routes not known to occur in humans.
Maternal diet
Maternal diet influences the FA composition of breast milk, with changes appearing within 8–10 h after a meal intake and contributing to approximately 30 % of total milk FA. The kind and levels of FA in breast milk can be modified by changes in dietary fat or energy intake. As demonstrated by Nasser et al. ( Reference Nasser, Stephen and Goh 27 ) in lactating women, breast milk medium-chain fatty acid (MCFA) concentrations were significantly higher when a low-fat, high-carbohydrate diet was consumed. These changes in MCFA concentrations suggest de novo FA synthesis from carbohydrates. In addition, incorporating fish into the diets of nursing mothers during lactation contributes to an increase of EPA, DPA and DHA in human milk( Reference Quinn and Kuzawa 35 ).
Trans-fatty acids (TFA) are FA found mainly in industrial partially hydrogenated oils and derived products (e.g. margarines, spreads, baked goods and fast foods). They are also found in meat and milk from ruminant animals (coming from production by bacterial metabolism of PUFA in the rumen)( Reference Stender, Astrup and Dyerberg 36 ). Maternal TFA intake is also reflected in breast milk( Reference Friesen and Innis 37 ).
Mobilisation of maternal body stores
The composition of human milk can be affected by the maternal nutritional status as well( 38 ). In cases of severe energy restriction, body fat is mobilised, for energy, and the composition of breast milk resembles that of maternal body fat stores. We found that, in lactating women who habitually ate a low-fat diet, the LA secreted in milk was not derived from direct intestinal absorption, suggesting that an important proportion of LA came from the maternal body pool. Turnover of maternal body stores seems to be the major source of milk LA in these women( Reference Del Prado, Villalpando and Elizondo 39 ).Conversely, recent studies realised in obese rats have demonstrated that the combination of maternal obesity and increased fat intake during gestation and lactation induces alterations in maternal liver metabolism( Reference Bautista, Montaño and Ramirez 40 ) and adversely changes maternal milk fat concentration( Reference Saben, Bales and Jackman 41 ). For example, Panagos et al.( Reference Panagos, Vishwanathan and Penfield-Cyr 42 ) reported that the breast milk from obese mothers had a higher n-6 to n-3 FA ratio and lower concentrations of DHA and EPA compared with lean mothers.
Parity has also been found to have an important effect on body fat stores. Previous studies have shown that maternal multi-parity may place offspring at a greater risk of decreased accretion of brain DHA if the maternal diet contains insufficient n-3 LC-PUFA( Reference Ozias, Carlson and Levant 43 ). This may be explained by the depletion of maternal DHA stores.
Nutrigenomics from milk fatty acids
Recent studies in mammals such as rodents, cows and humans have shown that lipids can regulate gene expression in liver and mammary gland, contributing to maintaining adequate concentrations of SFA, MUFA and PUFA in those tissues( Reference Ibeagha-Awemu, Li and Ammah 44 , Reference Rodriguez-Cruz, Sánchez and Bernabe-Garcia 50 ). Dietary lipids may act as regulators of lipogenesis interacting with transcription factors including the nuclear receptors PPAR and the transcription factors sterol-regulatory element binding protein (SREBP)( Reference Capel, Rolland-Valognes and Dacquet 45 , Reference Neschen, Morino and Dong 46 ). Both transcription factors are involved in the regulation of the genes FADS1 and FADS2 (encoding for the enzymes Δ5 and Δ6 desaturases, respectively) and the genes ELOV-2 and ELOV-5 (encoding for the enzymes elongases). PPAR comprise a superfamily including PPARα, PPARγ and PPARβ/δ. Activation of PPARγ and PPARβ/δ upregulates the expression of genes involved in de novo FA synthesis, whereas activation of PPARα upregulates genes that control FA oxidation( Reference Jump, Tripathy and Depner 47 ). SREBP are a family of transcription factors that have been characterised as mediators of cellular cholesterol homoeostasis and as regulators of FA biosynthesis and uptake. Three members of the SREBP family, SREBP-1a, SREBP-1c and SREBP-2, have been identified. Although SREBP-1c and SREBP-2 are structurally similar, their regulation in the liver by hormones, nutrients( Reference Jump, Tripathy and Depner 47 ) and during postnatal development is quite different( Reference Rodriguez-Cruz, Tovar and Palacios-Gonzalez 48 ). It has been discovered that whereas SREBP-2 favours cholesterol synthesis, SREBP-1a and SREBP-1c control FA synthesis, regulating genes including lipoprotein lipase, acetyl-CoA carboxylase α, FAS, SCD, FADS1 and FADS2, and FA ELOVL-2 and ELOVL-5 ( Reference Rodriguez-Cruz, Tovar and Palacios-Gonzalez 48 , Reference Ito, Nagasawa and Omae 49 ). SREBP-1c is synthesised as a larger precursor protein that is anchored to the endoplasmic reticulum. After proteolytic cleavage, the N-terminal domain migrates to the nucleus and activates target genes by binding to SRE( Reference Jump, Tripathy and Depner 47 ).
PUFA and their metabolites are the main FA that act at the level of the nucleus in conjunction with these transcription factors to regulate the lipogenic genes mentioned above( Reference Jump, Tripathy and Depner 47 ). This occurs mainly when there is a demand for FA – for instance, in pregnancy and lactation( Reference Rodriguez-Cruz, Sánchez and Bernabe-Garcia 50 , Reference Rodriguez-Cruz, Sánchez and Sánchez 51 ). PUFA reduce the nuclear content of SREBP-1a and 1c by accelerating the decay of SREBP-1c mRNA, therefore lowering the hepatic content of SREBP-1c mRNA and the synthesis of SREBP-1c precursor( Reference Takeuchi, Yahagi and Izumida 52 ). At the proteolytic processing level, PUFA decrease the mRNA transcription through lowering SREBP-1 binding to SRE on the promoter region of different lipogenic genes such as desaturases and elongases. Interestingly, SRE have been identified in the promoter region of FADS2 ( Reference Nara, He and Tang 53 ), ELOVL-5 and ELOVL-2 ( Reference Shikama, Shinozaki and Takeuchi 54 , Reference Kumadaki, Matsuzaka and Kato 55 ), although this has not been clearly demonstrated in FADS1. It is well recognised that, in liver and extrahepatic tissues that provide LC-PUFA for milk synthesis, the FADS1, FADS2, ELOVL-2 and ELOVL-5 genes are regulated by SREBP-1c through dietary lipids( Reference Rodriguez-Cruz, Tovar and Palacios-Gonzalez 48 ). This is consistent with the report by Tu et al. ( Reference Tu, Cook-Johnson and James 56 ), who found in rats that a diet containing very low levels of PUFA resulted in elevated hepatic mRNA expressions of FADS2 and ELOVL-2 genes relative to a diet richer in PUFA.
Our group identified for the first time that SREBP-1c is expressed in adipose tissue and mammary gland( Reference Rodriguez-Cruz, Tovar and Palacios-Gonzalez 48 ). In addition, Rudolph et al. ( Reference Rudolph, Neville and Anderson 57 ) found that, in pregnancy, SREBP-1a is expressed at a level about equal to SREBP-1c. However, only SREBP1-c is increased at the onset of lactation. On the other hand, we also found that lactating rats fed a low-PUFA diet (2 % maize oil rich in LA) had, in comparison with those fed a high-PUFA diet (10 % maize oil rich in LA), increased expression of SREBP-1c and its target genes FADS1 and FADS2 in mammary tissue and liver. At the same time, there was a reduction in the hepatic expression of PPARα, reducing the oxidation of FA. This was consistent with a higher (1–13C) LA incorporation by the mammary gland in the rats fed the low- compared with the high-lipid diet (26·66 (sd 4·77) v. 12·33 (sd 3·44) %, respectively) and a higher conversion of the uptaken LA into AA (1·06 (sd 0·43) v. 0·23 (sd 0·07) %, respectively)( Reference Rodriguez-Cruz, Sánchez and Bernabe-Garcia 50 ). In a recent study, we found that FADS1 and FADS2 mRNA were significantly overexpressed on certain days of pregnancy and lactation in the mammary gland, liver and adipose tissue from rats fed a low-lipid diet compared with rats fed an adequate-lipid diet. These results suggest that desaturase- and elongase-mediated LC-PUFA synthesis during pregnancy and lactation is not specific to the liver. The increased mRNA expression of these and other enzymes (FAS and SREBP-1c) in the mammary tissue involved in FA synthesis and regulation suggests an important role of this tissue, especially in mothers deficient in dietary lipids( Reference Rodriguez-Cruz, Sánchez and Sánchez 51 ). Additional research is necessary to determine whether other key regulators of lipogenesis in the lactating mammary gland exist, such as for example the carbohydrate response element binding protein (CHREBP). CHREBP may be indeed another regulator of lipogenesis in the mammary gland, as a high intake of the trans-10, cis-12 isomer of conjugated linoleic acid (18 : 2 trans-10, cis-12) has been shown to reduce CHREBP mRNA expression, as well as lipogenesis( Reference Harvatine, Boisclair and Bauman 58 ).
The effects of maternal TFA intake on the FA composition of breast milk differ depending on the type of isomer. Industrially produced TFA intake affects the milk FA composition negatively, and this occurs through various mechanisms: effects on gene expression or inhibition of the desaturation of LA and ALA to AA and DHA, respectively, in the mammary gland, with potential adverse effects on infant health( Reference Kadegowda, Connor and Teter 59 − Reference Innis 61 ). Previous studies have demonstrated that an excessive CLA and VA intake may reduce the milk’s fat density. This is explained because these ruminant-produced TFA are able to inhibit the expression of lipogenic genes related to FA synthesis (ACACA, FAS), FA desaturation (SCD1, SCD2), TAG esterification (AGPAT1) and transcriptional factors (SREBP-1c and CHREBP)( Reference Kadegowda, Connor and Teter 59 ); this process has been called milk fat depression (Fig. 2).
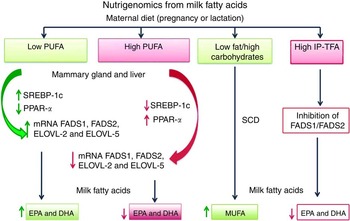
Fig. 2 Nutrigenomics from milk fatty acids. The effect of low or high amount of PUFA and high quantity of industrially produced trans-fatty acids (IP-TFA) from maternal diet on fatty acid desaturases (FADS1, FADS2), elongases (ELOVL-2, ELOVL-5), sterol-regulatory element binding protein-1 (SREBP-1c), stearoyl-CoA desaturase (SCD) and PPARα mRNA expression levels in liver and mammary gland during the pregnancy or lactation.
The scientific evidence presented above suggests that maternal dietary FA partly determine the FA composition of human milk by modulating gene expression of key transcription factors such as SREBPs, PPAR and CHREB, among others. During lactation, these transcription factors regulate FA synthesis and oxidation; therefore, those mechanisms are triggered to face the demands of the newborn for development and growth.
Nutrigenetics from milk fatty acids
The impact of genetic variation on milk FA composition has been (and continues to be) widely explored in cows and other ruminant animals. Quantitative trait loci mapping studies have helped to identify genomic regions associated with milk fat composition and to select candidate genes within these regions for further study( Reference Schennink, Stoop and Visker 62 , Reference Stoop, Schennink and Visker 63 ). Candidate gene studies have indeed shown that SNP in selected genes are associated with the FA composition of milk( Reference Mele, Conte and Castiglioni 64 − Reference Tăbăran, Balteanu and Gal 70 ). The goal behind such studies has been generating the knowledge needed to be able to use selective breeding (together with nutritional manipulation) to breed animals that produce milk with an ‘improved’ FA composition. ‘Improved’ composition in this context means a composition that is healthier for humans on the one hand and able to satisfy practical consumer demands on the other (e.g. butter with better spreadability). Nowadays, the focus lays on enhancing the content in milk of C18 MUFA and that of PUFA, particularly EPA, DHA and CLA( Reference Ashes, Gulati and Scott 71 − Reference Lanier and Corl 73 ).
Studies on the impact of genetic variation on human milk FA composition are instead scarce. They probably only started after the first associations between SNPs and proportions of FA (namely, LC-PUFA) in blood and/or tissues had been reported. Schaeffer et al. ( Reference Schaeffer, Gohlke and Müller 74 ) were the first ones to show that common SNPs in the genes coding for FADS were associated with percentages of LC-PUFA in plasma phospholipids. These results were later confirmed by others( Reference Malerba, Schaeffer and Xumerle 75 − Reference Moltó-Puigmartí, Plat and Mensink 81 ). At the same time, Rodriguez-Cruz et al.( Reference Rodriguez-Cruz, Tovar and Palacios-Gonzalez 48 ) had demonstrated that fatty acid desaturases were present in the rat mammary gland, raising the question as to whether desaturases were also present in the human mammary gland for the synthesis of LC-PUFA to be secreted into breast milk. Triggered by these findings, Xie & Innis( Reference Xie and Innis 80 ) and Moltó-Puigmartí et al.( Reference Moltó-Puigmartí, Plat and Mensink 81 ) investigated and demonstrated that FADS SNP were not only associated with LC-PUFA levels in human plasma as Schaeffer et al. ( Reference Schaeffer, Gohlke and Müller 74 ) had shown but also with LC-PUFA levels in breast milk( Reference Xie and Innis 80 , Reference Moltó-Puigmartí, Plat and Mensink 81 ). Namely, women homozygous for the minor alleles had lower AA and DHA proportions in their plasma and breast milk than women carrying the major allele. Moreover, Moltó-Puigmartí et al. ( Reference Moltó-Puigmartí, Plat and Mensink 81 ) found that FADS SNP modified the effect of fish intake on breast milk DHA levels; concretely, DHA increased with DHA (i.e. fish) intake in women carrying the major allele but not in women homozygous for the minor allele, at least up to the observed intake level (Fig. 3)( Reference Moltó-Puigmartí, Plat and Mensink 81 ). This finding is of public health interest given that the supply of LC-PUFA to the child during lactation is key for a proper (neuro)development, and breast-fed children from women homozygous for the minor allele could be therefore in a certain disadvantage compared with children from women carrying the major allele. In the years following these first findings, three additional studies have confirmed the association of FADS SNP with breast milk FA( Reference Morales, Bustamante and González 82 – Reference Ding, Liu and Li 84 ). Moreover, two studies have shown that also SNPs on the genes coding for the enzymes elongases (namely, ELOVL-2 and ELOVL-5) are associated with the proportions of PUFA in breast milk( Reference Morales, Bustamante and González 82 , Reference Li, Gan and Ding 85 ). It is to be expected that, with the development of genotyping techniques and the knowledge on FA metabolism, new genetic variants associated with breast milk FA composition will be discovered in the near future. To the best of our knowledge, replication of the gene–diet interaction described by Moltó-Puigmartí et al. has not yet been attempted. However, such an interaction illustrates the possibility that genetic variants contributing to the variability of human milk (FA) composition interplay with maternal diet, bringing the opportunity to modify (i.e. improve) the composition of breast milk through nutrigenetics.

Fig. 3 Breast milk DHA proportions as a function of maternal fatty fish and fish oil intake. Regression lines for major allele homozygous women are represented.
Discussion and conclusions
Human breast milk is an optimal food for infants because it contains a wide variety of nutrients and bioactive molecules that contribute to healthy growth and (neuro)development. Several studies have shown that maternal nutrition during pregnancy and lactation affects milk composition and hence influences infant’s development. In this paper, we review the role of nutrigenomics and nutrigenetics on breast milk FA composition. On the one hand, FA from maternal diet are able to interact directly or indirectly with some transcription factors including the nuclear receptor PPAR, SREBP and CHREBP. This interaction produces changes in the composition of milk. For example, SREBP and its target genes FADS1, FADS2, ELOVL-2 and ELOVL-5 are regulated by the maternal intake of PUFA to maintain adequate concentrations of PUFA in human milk. Instead, industrially produced TFA intake affects the milk FA composition negatively and may have adverse effects on growth and development by interfering with essential FA metabolism. On the other hand, SNP in the FADS and ELOVL genes are associated with PUFA levels in breast milk, and may affect the maternal response to FA intake.
Acknowledgements
This research received no specific grant from any funding agency, commercial or not-for-profit sectors.
M. R.-C. was in charge of the interpretation of the articles reviewed and manuscript design and editing. E. S.-C. and C. M.-P. had responsibility for research information and manuscript writing. All authors contributed to discuss and had input writing the article. All authors contributed equally to the literature search, analysis of the data published, manuscript writing and revisions of the article. All authors approved the final version of the manuscript.
The authors declare that there are no conflicts of interest.





