The advent of the molecular characterisation of intestinal microbiota has occasioned an increased interest in the relationship between diet, microbiota and nutrition. Unabsorbed dietary carbohydrates form the major energy source for the colonic microbiota. In turn, the bacteria ferment these to SCFA which are absorbed and enter the portal venous system, and contribute to energy in the host. Changes in gut microbiota have been described in association with obesity and have been linked to increased energy harvest(Reference Turnbaugh and Gordon1–Reference Balamurugan, George and Kabeerdoss3). Vegetarian diets have been promoted for specific conditions including induction of weight loss and improvement in insulin resistance(Reference Thedford and Raj4, Reference Kahleova, Matoulek and Malinska5). The influence of a vegetarian diet on faecal microbiota remains speculative. In an early study based on quantitative culture, consumption of meat increased the counts of Bacteroides, Bifidobacterium, Peptococcus and anaerobic Lactobacillus species(Reference Reddy, Weisburger and Wynder6). On the other hand, a recent study using molecular techniques has reported that young adults had a predominance of Clostridium cluster IV bacteria in faecal microbiota compared with elderly individuals, with omnivores among the young adults having marginally higher proportions of these bacteria than vegetarians(Reference Liszt, Zwielehner and Handschur7).
Rural populations in southern India consume a largely vegetarian diet. There are two factors that determine whether these populations eat any meat. The first relates to socio-economic circumstances as a result of which meat forms a part of the diet only once or twice per week. The second is religious persuasion leading to total avoidance of meat intake in significant sections of these populations. These dietary patterns are most distinct among women in these populations. The present study examined a cohort of young women from a rural background studying in a women's college in southern India, and reports the dietary intakes, nutritional status and faecal microbiota of this cohort, with particular reference to the differences between omnivores and vegetarians.
Methods
A total of thirty-two vegetarian and twenty-four non-vegetarian young women (aged 18–27 years), all from a very similar socio-economic background, were recruited from resident students in a women's college in Tamilnadu. The study was preceded by focus group discussions in the college with the teachers and students where the purpose of the study was explained and students were invited to participate. Participants were excluded if they had consumed antibiotics within the last 1 month or had gastrointestinal symptoms at the time of the study. A 24 h diet recall, together with an FFQ of commonly used foods ingested over the previous 3 months, was administered by a trained dietitian. Food quantities were measured according to a standard set of cups and spoons, and the daily intake of relevant nutrients was calculated from standard tables for the composition of Indian foods(Reference Gopalan, Rama Sastri and Balasubramanian8). Height and weight were measured and BMI was calculated. The socio-economic score was calculated according to the method of Kuppuswami modified for 2007(Reference Kumar, Shekhar and Kumar9). Each participant provided a sample of freshly passed stool in the morning, and this was transported to the laboratory on ice within 1 h and stored in aliquots at − 80°C until DNA extraction. The study was conducted according to the guidelines laid down in the Declaration of Helsinki and all procedures involving human subjects/patients were approved by the Institutional Review Board of the Christian Medical College, Vellore. Written informed consent was obtained from all participants.
Faecal DNA was extracted using QiaAmp DNA Stool Mini Kits (Qiagen). Faecal bacteria were analysed by quantitative PCR using primers targeted at 16S ribosomal DNA of Clostridium coccoides–Eubacterium rectale group (Clostridium cluster XIVa)(Reference Matsuki, Watanabe and Fujimoto10), Clostridium leptum group (Clostridium cluster IV)(Reference Matsuki, Watanabe and Fujimoto11), Faecalibacterium prausnitzii (Reference Balamurugan, Janardhan and George12), Desulfovibrio species(Reference Balamurugan, Janardhan and George12), Bacteroides–Prevotella–Porphyromonas group(Reference Balamurugan, Janardhan and George12), Enterococcus faecalis (Reference Balamurugan, Janardhan and George12), Bifidobacterium genus(Reference Sokol, Pigneur and Watterlot13), Lactobacillus group(Reference Sokol, Pigneur and Watterlot13), Enterobacteria(Reference Ahmed, Macfarlane and Fite14), Ruminococcus productus–C. coccoides (Reference Malinen, Rinttilä and Kajander15), Butyrivibrio fibrisolvens (Reference Balamurugan, Aarthi and Chittaranjan16), and Roseburia spp.–E. rectale (Reference Ramirez-Farias, Slezak and Fuller17). The latter are the principal butyrate-producing bacteria within Clostridium cluster XIVa. Together with F. prausnitzii that belongs to Clostridium cluster IV, they form the major butyrate-producing bacteria in the intestine. We were particularly interested in studying butyrate-producing bacteria, and quantified the butyryl-CoA CoA-transferase gene in the faeces using BCoATscrF and BCoATscrR primers(Reference Louis and Flint18). The specificities of these primers and the PCR conditions have been reported earlier. Lactobacillus group primers detected Lactobacillus, Pediococcus and Leuconostoc group bacteria. The microbial groups chosen for the study represented numerically or biologically important constituent classes of faecal bacteria. Real-time PCR was performed in a Chromo4 system (Bio-Rad Laboratories) using the Mesa Green quantitative PCR Mastermix Plus for SYBR Assay (Eurogentec)(Reference Balamurugan, Janardhan and George12). 16S ribosomal (‘universal’) DNA sequences conserved in Domain Bacteria were amplified simultaneously using appropriate primers(Reference Suzuki, Taylor and DeLong19) and served as the denominator, against which quantitative amplification of the other 16S ribosomal DNA amplicons was reported. DNA copy was expressed as the ‘relative difference’, i.e. the cycle threshold at which DNA for each target was detected relative to the cycle threshold at which Domain Bacteria DNA was detected upon amplification. The reference universal amplicon was assigned a value of 1, and target microbial groups are expressed as decimal values in proportion to this.
Statistics
All values are presented as medians and interquartile ranges. Comparisons between the groups were done by t tests using log-transformed values for the relative difference. Log-transformed values of the transferase gene and of the microbial communities were used to examine their correlations (Pearson) with dietary components and with one another, as well as for linear regression. Two-tailed P values less than 0·05 were considered to be statistically significant.
Results
In the present study, thirty-two vegetarians and twenty-four omnivores were enrolled in the study. There were no significant differences in age, height, weight, BMI or socio-economic score between the two groups (Table 1). Total energy intake, complex carbohydrates and Ca were significantly higher in the omnivorous group than in the vegetarian group (Table 1).
Table 1 Demographics, anthropometric data and dietary intakes of the study participants
(Median values and interquartile ranges)

* P values from Mann–Whitney U tests for the assessment of significant differences between the median values.
Faecal levels of the C. coccoides–E. rectale group (Clostridium cluster XIVa bacteria) were higher in the omnivores (relative difference: median 0·08 108, interquartile range 0·04 229–0·1247) compared with the vegetarian group (median 0·03 158, interquartile range 0·00 912–0·06 229; P = 0·004). This was contributed by significant increases in Roseburia–E. rectale (P = 0·023), whereas R. productus–C. coccoides and Butyrivibrio were not increased (Fig. 1). All the other microbial groups tested were similar between the two groups (Fig. 1). Interestingly, butyryl-CoA CoA-transferase gene levels were significantly higher in the omnivore group compared with the vegetarian group (P = 0·019, Fig. 1). Log-transformed levels of the gene were significantly correlated with C. coccoides–E. rectale (Pearson r 0·607, P < 0·0001) and Roseburia–E. rectale (Pearson r 0·777, P < 0·0001) but not with R. productus–C. coccoides. Transferase gene levels were negatively correlated with fat intake (r − 0·288, P = 0·032) and showed a trend for a positive correlation with crude fibre intake (r 0·231, P = 0·089). C. coccoides–E. rectale and R. productus–C. coccoides abundance correlated with crude fibre intake (r 0·33, P = 0·014; r 0·30, P = 0·022, respectively). Linear regression with butyryl-CoA CoA-transferase as the dependent variable and various microbial communities, crude fibre and fat as independent variables showed that Roseburia–E. rectale (P = 0·000) and C. coccoides–E. rectale (P = 0·044) were the only variables that independently correlated with log abundance of the transferase gene.
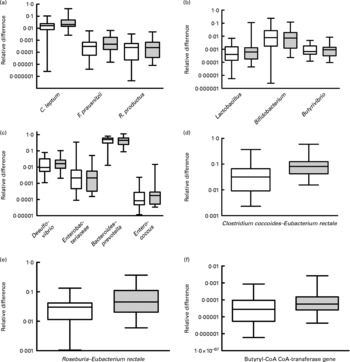
Fig. 1 Composite figure showing the individual groups, genera or species quantified by quantitative PCR (qPCR) with SYBR Green and primers targeting 16S rRNA gene sequences specific for the microbial communities of interest. Specificity of amplification was examined by amplicon size as well as melting curve analysis. The y-axis is logarithmic and the box and whisker plots show the medians, interquartiles and total ranges of the relative difference of each group relative to the universal Domain Bacteria sequence that was amplified in parallel with the same qPCR. (d) Values were significantly different (P = 0·008). (e) Values were significantly different (P = 0·037). (f) Values were significantly different (P = 0·045). □, Vegetarians; ![]() , omnivores.
, omnivores.
Discussion
The present study evaluated differences in faecal microbiota that could be related to a vegetarian or omnivore diet in a well-matched population of young women in southern India, and found increases in Clostridium cluster XIVa bacteria in the omnivore group compared with the vegetarian group. This was accompanied by an increase in the genes coding for a key enzyme involved in butyrate production.
The two groups of participants were very well matched, being within a narrow age range, of the same sex, all from the same geographical area and all accustomed to food prepared in a typical southern Indian style. The vegetarians in this group consumed small amounts of cows' milk on a daily basis, in keeping with traditional practices in southern India, and were not vegans. Vegetarian diets are associated with lower BMI, lower overall cancer rates and a lower risk of death from IHD(Reference Craig and Mangels20). In the present study, BMI of the participants in the vegetarian group did not significantly differ from that of the participants in the omnivorous group. The intake of Ca, normally quite low among women in India, was particularly low in the vegetarian group compared with the omnivore group.
Dietary preferences play a major role in shaping the intestinal microbiota. Unabsorbed dietary carbohydrate, consisting of amylase-resistant starch and NSP, is the major energy source for the microbiota. The latter, in turn, helps the host by salvaging otherwise ‘lost’ energy through fermentation of unabsorbed carbohydrate and production of SCFA which contribute to energy in the human host. There are only a few studies of the faecal microbiota of vegetarians in the molecular era. A recent study has used molecular (PCR-denaturing gradient gel electrophoresis) analysis of the faecal microbiota of vegetarians and omnivores but failed to show significant differences in faecal microbiota; however, the authors noted a tendency for Clostridium cluster IV bacteria and F. prausnitzii (belonging to the same cluster) to be found more frequently in the faeces of omnivores compared with vegetarians(Reference Liszt, Zwielehner and Handschur7).
In the present study, the faecal microbiota contained significantly greater numbers of the C. coccoides–E. rectale group (belonging to Clostridium cluster XIVa) in omnivorous women compared with vegetarian women. Bacteria belonging to Clostridium clusters XIVa and IV are noted for their ability to ferment carbohydrate to SCFA. SCFA, especially butyrate, have physiologically important effects on the colonic epithelium and on host metabolism. Dominant butyrate-producing bacteria including Roseburia species, E. rectale, Ruminococcus species and Butyrivibrio species(Reference Louis, Young and Holtrop21) belong to Clostridium cluster XIVa. Using appropriate primer pairs, we have shown that the faecal microbiota of omnivores was enriched with Roseburia–E. rectale compared with vegetarians, whereas R. productus–C. coccoides and Butyrivibrio were not significantly different between the two groups. Butyryl-CoA CoA-transferase is an enzyme involved in butyrate synthesis by the microbiota(Reference Louis and Flint18, Reference Louis, Young and Holtrop21), and its gene levels correlated with the levels of both the Roseburia–E. rectale group and cluster XIVa bacteria as a whole, possibly indicating that these microbial groups are the predominant butyrate-producing bacteria in this population. Quantitative PCR was the only tool used to assess the abundance of subgroups and showed an increased relative abundance of Clostridium cluster XIVa bacteria (C. coccoides–E. rectale) generally and specifically of the Roseburia–E. rectale group within this cluster. Although differences in the abundance of the subgroups may be compensated by other microbial groups that provide the same metabolic capacity, we also found a difference in metabolic capacity (i.e. in butyryl-CoA CoA-transferase gene numbers) for producing butyrate, which suggests that the difference in subgroups between vegetarians and omnivores is relevant. The reason for the increased abundance of Roseburia–E. rectale in omnivores was not clear. Although crude fibre intake and fat intake correlated with the abundance of specific microbial communities in the group as a whole, they did not explain the differences between vegetarians and omnivores. It is possible that dietary constituents not measured in the study or complex nutrient–microbiota–microbiota interactions are responsible for this observation.
In conclusion, comparison of the faecal microbiota of lacto-vegetarian and omnivorous young women showed that Clostridium cluster XIVa bacteria and butyrate-producing bacteria were significantly more abundant in the faecal microbiota of omnivores. Identifying the reason for this phenomenon may require more detailed approaches involving metagenomic and metabonomic analysis.
Acknowledgements
J. K. was supported by a Senior Research Fellowship from the Indian Council of Medical Research. This study was supported by grants from the Department of Biotechnology, New Delhi. J. K. was responsible for the conception, design, sample analysis, data analysis and writing of the manuscript; R. S. D. was responsible for participant recruitment and dietary analyses; R. R. M. was responsible for conception and participant recruitment; B. S. R. was responsible for the conception, design and writing of the manuscript. The authors have no conflicts of interest to report.




