A better understanding of ethanol on diseases and sub-health conditions will have a huge social and economic impact on our life. Epidemiological and controlled clinical studies have investigated the effects of varying levels and patterns of alcohol consumption on type 2 diabetes(Reference Flanagan, Moore and Godsland1–Reference Carlsson, Hammar and Grill3). A U-shaped relationship has been uncovered that means moderate ethanol may protect against diabetes, while excessive ethanol is a risk factor for type 2 diabetes(Reference Koppes, Dekker and Hendriks4). However, the exact mechanisms underlining the diabetogenic effects of ethanol have not been fully expatiated.
Type 2 diabetes manifests itself in individuals when the β-cell fails to compensate for peripheral insulin resistance to maintain normoglycaemia and evidence from man and rats suggests that this may be in part because of a decreased β-cell mass(Reference Leahy5–Reference Pick, Clark and Kubstrup7). Furthermore, much evidence suggests that insulin resistance is necessary but not sufficient for the development of type 2 diabetes(Reference Weyer, Bogardus and Mott8–Reference Ferrannini, Gastaldelli and Miyazaki10), and reduced β-cell mass plays a pivotal role in determining whether an individual will progress to type 2 diabetes(Reference Ahrén11–Reference Rhodes13). The effect of ethanol on β-cell mass has not been reported yet.
Muscle and adipose tissue are the two major sites of glucose disposal in response to insulin. The impairments of the insulin signalling pathway in these two tissues appear to be critical to peripheral insulin resistance. Insulin signalling is a complex and highly integrated network that mediates the physiological functions of insulin. The phosphatidylinositol 3-kinase (PI3K) pathway is responsible for most of the metabolic actions of insulin. Briefly, insulin binding to its receptor results in the phosphorylation of insulin receptor substrates (IRS). Phosphorylated IRS proteins then associate with PI3K through the p85 regulatory domain leading to the activation of PI3K. The activation of PI3K leads to the downstream phosphorylation of Akt/protein kinase B and atypical protein kinase B, which can ultimately result in the translocation of GLUT-4 and glucose uptake(Reference Chang, Chiang and Saltiel14–Reference Cantley16). Clearly, PI3K and GLUT-4 play central roles in the insulin-stimulated glucose disposal process(Reference Taniguchi, Emanuelli and Kahn17).
Though the mechanisms underlining the conditional diabetogenic effects of ethanol have not been clearly expatiated, insulin resistance in peripheral insulin target organs and reduced β-cell mass are two practical indexes to follow. Although conditional ethanol effects on insulin sensitivity are well established(Reference Tomie Furuya, Binsack and Onishi18, Reference Onishi, Honda and Ogihara19), studies of ethanol effects on β-cell mass are rare. To our knowledge, ethanol effects on both insulin resistance and β-cell mass have not been studied. In the present report, the impacts of ethanol on insulin resistance and β-cell mass were investigated at the physiological and molecular levels.
Materials and methods
Materials
Wistar rats (153 (sd 8) g body weight) were purchase from the Laboratory Animal Center of Hubei Provincial Center for Disease Control and Prevention, Hubei, China. Human recombinant insulin was obtained from Novo Nordisk. 125I-Insulin radioimmunoassay kit was purchased from the Isotope Institute of China Institute of Atomic Energy. Trizol reagent was obtained from Invitrogen. SYBR® Premix Ex Taq™ (Perfect Real Time) was from TaKaRa Bio Inc., ATP was purchased from Upstate Biotechnology, protein A-Sepharose and chemiluminescence reagents (ECL kit) were obtained from Amersham Pharmacia Biotech, nitrocellulose membranes were from Sigma, protein assay kits were purchased from Nan Jing Jian Cheng Bio Inc., α-phosphatidylinositol was purchased from Calbiochem Corp. and [γ-32P]ATP was obtained from Rui Fu Biotechnology Company (Beijing, China). Antibodies used in the experiment were obtained from the following sources: purified rabbit anti-IRS-1 antibody (Upstate Biotechnology); purified goat polyclonal anti-Glut-4 antibody and rabbit anti-insulin antibody (Santa Cruz, USA); monoclonal mouse anti-β-actin antibody, horseradish peroxidase-labelled anti-goat IgG, horseradish peroxidase-labelled anti-mouse IgG and biotin goat anti-rabbit IgG (Sigma).
Animals
All the experimental protocols used in the present study were approved by the Animal Care and Use Committee of Hua Zhong University of Science and Technology. Wistar rats were housed at 17–27°C, 40–60 % relative humidity with a 12:12 h dark–light schedule. To initiate the study, forty animals were weighed and randomly assigned to four testing groups and subjected to daily intragastric injections (10 ml/kg body weight per d) containing different levels (0, 10, 20, 33 % v/v) of ethanol for 19 weeks. Animals were maintained on normal rodent chow.
Animal characterization
Body weight was monitored daily during the feeding period. For fasting plasma glucose and fasting plasma insulin, blood samples were collected after 12 h of starvation and immediately subjected to the assays. Blood glucose levels were measured by the glucose oxidase method and serum insulin was determined by radioimmunoassay.
Intraperitoneal insulin tolerance test
Intraperitoneal insulin tolerance test was done as described previously(Reference Sridhar, Vinayagamoorthi and Arul Suyambunathan20, Reference Yuan, Konstantopoulos and Lee21). Briefly, after 6 h of fasting, insulin (2·0 units/kg) was injected intraperitoneally into the testing rats (four groups) and blood samples were collected at different time intervals for plasma glucose by glucose. Area under the curve was calculated.
Insulin stimulation
Insulin stimulation was performed by intraperitoneal injection of 15 units human recombinant insulin/kg into five rats from each testing group. The other rats were used as the matched non-insulin control. The animals were killed 30 min after the injection, and the gastrocnemius muscle and perirenal adipose tissues of the rats were collected, frozen in liquid N2 and stored at − 70°C for subsequent molecular assays.
Pancreas histology
After the animals were killed, pancreatic specimens were harvested for histological study. Specimens were fixed in a 10 % formalin solution for 120 h and then set in paraffin blocks for section. Three representative glass slices of 6 μm thick sections were randomly obtained from each animal and stained with haematoxylin–eosin.
Immunohistochemistry and morphometric analysis
Immunohistochemistry and morphometric analysis of the rat pancreatic islet were employed as described(Reference Elayat, el-Naggar and Tahir22). Rat pancreas sections (6 μm thick) were regularly deparaffinaged. After washing in PBS, slides were blocked at room temperature with 3 % H2O2 followed by 10 % normal goat serum for 30 min. The treated samples were then sequentially incubated with rabbit anti-insulin antibody (overnight at 4°C), biotin goat anti-rabbit IgG (30 min, room temperature), spereptavidin–biotin–peroxidase complex (30 min, room temperature) and 3,3-diamino-benzidinetetrahydrochloride solution (10 min, room and temperature). To appreciate the negative nuclei better, the slices were counterstained with haematoxylin. Islet texture characteristics were analysed by the HPIAS-1000 image analytic system (Ins+/panc is the volume density of the insulin immunoreactivity per pancreatic tissue; Ins+/Isl is the volume density of the insulin immunoreactivity per islet).
Insulin receptor substrate-1-associated phosphatidylinositol 3-kinase activity
The IRS-1 associated PI3-K activity was determined as described(Reference Barbour, Shao and Qiao23). Muscle and adipose tissue were collected and homogenized(Reference Wojtaszewski, Hansen and Ursø24) in ice-cold buffer (50 mm-HEPES, 137 mm-NaCl, 10 mm-Na4P2O7, 10 mm-NaF, 1 mm-MgCl2, 1 mm-CaCl2, 1 % Nonidet P-40, 10 % glycerol, 2 mm NaVO4, 2 g/ml aprotinin, 5 g/ml leupeptin, 0·5 g/ml pepstatin, 10 g/ml antipain, 1·5 mg/ml benzamidine and 100 μmol/l 4-(-2-aminoethyl)benzenesulphony fluoride hydrochloride and rotated for 1 h at 4°C. Homogenates were centrifuged at 4°C at 12 000 g for 15 min. Supernatant was collected and protein content was estimated by the Bradford method. Muscle and adipose homogenates (100 mg protein) were immunoprecipitated with IRS-1 antibody overnight at 4°C (400 g protein/4 g antibody), followed by incubation with protein A-Sepharose overnight. The immunoprecipitation complex was spun at 14 000 g for 10 min, followed by washing three times with isotonic PBS containing 1 % Nonidet P-40, twice in 0·5 m-LiCl2/100 mm-2-amino-2-hydroxymethyl-1,3-propanediol (Tris)-HCl (pH 7·6), and twice with 10 mm-Tris-HCl (pH 7·4), 100 mm-NaCl and 1 mm-EDTA. The pellets were resuspended in 50 μl of the final wash buffer, and 10 μl 100 mm-MgCl2 were added along with 10 μl of a mixture containing 0·5 mg/l α-phosphatidylinositol in 10 mm-Tris/1 mm-EGTA, and the tube was sonicated for 20 s. To start the PI3K reaction, 10 μl ATP mix containing 100 mm-MgCl2, 10 mm-Tris (pH 7·5), 0·55 mm-ATP and 1 mCi/ml [γ-32P]ATP were added for 10 min at room temperature. The reaction was stopped with 20 μl 8 m-HCl for 5 min followed by 160 μl CHCl3–MeOH (1:1). The phases were separated by centrifugation, and the lower organic phase was removed. The aliquots were spotted on to TLC plates, resolved in CHCl3–MeOH–H2O–NH4OH (60:47:11·3:2), dried and visualized by autoradiography, and the phosphatidylinositol 3-phosphate spot was cut out and quantified in a scintillation counter. PI3K activity is expressed in arbitrary units relative to the control group.
Real-time PCR
Total RNA from tissues was isolated with Trizol agent. RNA concentration was determined by a spectrophotometer at A260 and designated the purity valid if the ratio of A260/A280 was in the range from 1·8 to 2·0. Using the RT system (Promega, Madison, WI, USA), cDNA was synthesized from 3 μg RNA at 37°C for 60 min followed by 95°C for 5 min. Real-time PCR was performed with SYBR® Premix Ex Taq™ (Perfect Real Time) using the ABI 7900HT real-time thermocycler (Applied Biosystems, Foster, CA, USA) with the following programme: 95°C/10 s (95°C/5 s, 60°C/30 s) × 40. The dissociation curve was performed and analysed using the ABI 7900HT software. Target gene was analysed and normalized to β-actin. The pairs of primers for GLUT-4 and β-actin reactions were 5′-TGG CAT GAT TTC CTC CTT TC-3′/5′-CAC CAA CCC TGA TGT TAG CC-3′ and 5′-CAT CAC TAT CGG CAA TGA GC-3′/5′-GAC AGC ACT GTG TTG GCA TA-3′, respectively. The CT value represents the number of cycles required for the fluorescence signal reach at threshold for each reaction (ΔCT = CT (target gene) − CT (β-actin)). The relative expression levels of GLUT-4 and β-actin were calculated as 2− ΔΔCT(Reference Livak and Schmittgen25).
Western blotting
An equal amount of tissue proteins was separated by SDS–PAGE and transferred to nitrocellulose membranes by electromembrane transfer for 2 h. The membranes were processed through the following steps: incubated in TBS-T (50 mm-Tris, 150 mm-NaCl, plus 0·1 % (v/v) Tween 20) with 3 % (w/v) BSA for 2 h, washed twice with TBS-T, incubated with a specific antibody in TBS-T with BSA (1 %) overnight at 4°C, washed again and probed with horseradish peroxidase-coupled goat anti-rabbit or anti-mouse IgG Fab fragments for 1 h. Immunoreactive bands were detected by enhanced chemiluminescence reagent (ECL kit) and the targeted protein was visualized by autoradiography. β-Actin was used as an internal control and the detected protein bands were quantified with a Biometra densitometer (Biometra, Goettingen, Germany).
Statistical analysis
SPSS version 12.0 software package (SPSS Inc., Chicago, IL, USA) was used for statistical analysis (SN: 59 245 46 841 40 655 89 389 09 859 21 671 21 957 29 589 12). Data were summarized as means and standard deviations. Statistical comparison of means among individual groups was done using a one-way ANOVA and two-sided Dunnett t test. Significant level was set at P < 0·05.
Results
Animal characterization
Body weight did not differ among groups at any time during the 19 weeks of the feeding period (Fig. 1). After 19 weeks of daily ethanol administration, the detected fasting plasma insulin levels in the 10 and 20 % (v/v) ethanol-loaded groups were significantly higher than the control (Table 1); whereas the fasting plasma glucose level of the 33 % (v/v) ethanol-loaded groups was significantly higher (18 %) than the control (Table 1).

Fig. 1 Body weight of rats. (![]() ), control group; (
), control group; (![]() ), 10 % (v/v) ethanol-loaded group; (
), 10 % (v/v) ethanol-loaded group; (![]() ), 20 % (v/v) ethanol-loaded group; (
), 20 % (v/v) ethanol-loaded group; (![]() ), 33 % (v/v) ethanol-loaded group. Values are means with their standard errors depicted by vertical bars (n 10). Body weight did not differ among groups at any time during the feeding period.
), 33 % (v/v) ethanol-loaded group. Values are means with their standard errors depicted by vertical bars (n 10). Body weight did not differ among groups at any time during the feeding period.
Table 1 Effect of chronic ethanol intake on fasting plasma glucose and fasting plasma insulin†
(Mean values and standard deviations for ten rats)
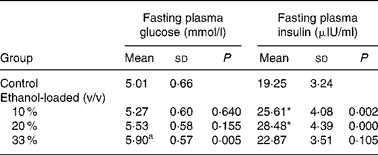
* Mean values were significantly different from those of the control group (two-sided Dunnett t test) (P < 0·05).
† For details of procedures, see Materials and methods.
Intraperitoneal insulin tolerance test
Insulin sensitivity was decreased in a dose-dependent manner (r − 0·842, P < 0·001) (Fig. 2) in three ethanol-loaded groups reflected by higher plasma glucose concentration after an intraperitoneal insulin injection.

Fig. 2 Rat intraperitoneal insulin tolerance test. After 6 h fast, rats were injected with insulin intraperitonealy (2·0 units/kg body weight) and blood samples were taken at different time intervals for plasma glucose level (a). (b), Area under the curve of different groups was calculated. Values are means with their standard errors depicted by vertical bars (n 10). (![]() ), control group (C); (
), control group (C); (![]() ), 10 % (v/v) ethanol-loaded group (L); (
), 10 % (v/v) ethanol-loaded group (L); (![]() ), 20 % (v/v) ethanol-loaded group (M); (
), 20 % (v/v) ethanol-loaded group (M); (![]() ), 33 % (v/v) ethanol-loaded group (H). *Mean values were significantly different from those of the control group (two-sided Dunnett t test (P < 0·05).
), 33 % (v/v) ethanol-loaded group (H). *Mean values were significantly different from those of the control group (two-sided Dunnett t test (P < 0·05).
Pancreas histopathology
Vacuolization and oedematous damage were detected in the pancreas collected from the 33 % (v/v) ethanol-loaded groups. Necrotic/haemorrhagic injury was also seen (Fig. 3).

Fig. 3 Histological evaluation of excessive ethanol-induced pancreatic injury in rats. Representative photomicrographs of rat pancreas stained by haematoxylin–eosin ( × 400) showed vacuolization and oedematous injury in pancreatic islet cells; necrotic/haemorrhagic injury was also seen in the pancreas. The results are representative of three separate experiments. C, control group; H, 33 % (v/v) ethanol-loaded group.
Immunohistochemistry and morphometric analysis
Insulin-positive area was distributed in rat islet (Fig. 4). The results of the morphometric analysis indicated that islet β-cell mass was significantly reduced in the 33 % (v/v) ethanol-loaded groups (Table 2).

Fig. 4 Insulin immunohistochemical staining of pancreas. Transversal sections of pancreas tissues ( × 400) from the control (C) and the 33 % (v/v) ethanol-loaded group (H) rats were immunolabelled with antibodies against insulin and biotinylated secondary antibodies as described in the Materials and methods. The results are representative of three separate experiments.
Table 2 Islet texture characteristics of rats†
(Mean values and standard deviations for ten rats)
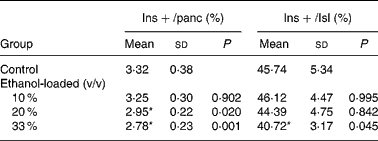
Ins+/Isl, the volume density of the insulin immunoreactivity per islet; Ins+/panc, the volume density of the insulin immunoreactivity per pancreatic tissue.
* Mean values were significantly different from those of the control group (two-sided Dunnett t test) (P < 0·05).
† For details of procedures, see Materials and methods.
Insulin receptor substrate-1-associated phosphatidylinositol 3-kinase activity
Insulin stimulated PI3K activity was decreased in a dose-dependent manner (r − 0·956, P < 0·001) in muscle tissue in all ethanol-loaded groups, whereas PI3K activity was significantly (P < 0·05) increased in adipose tissue of rats in 20 % (v/v) ethanol-loaded rats (Fig. 5).

Fig. 5 Insulin receptor substrate-1-associated phosphatidylinositol 3-kinase (PI3K) activity in muscle (a) and adipose (b) tissue of rats. Muscle and adipose extracts were obtained before and after in vivo insulin stimulation. PI3K activity was determined as described in the Materials and Methods. C, control group; L, 10 % (v/v) ethanol-loaded group; M, 20 % (v/v) ethanol-loaded group; H, 33 % (v/v) ethanol-loaded group. PI3K activity is expressed in arbitrary units relative to the control groups. Values are the average PI3K activity of three independent experiments without ( − ) and with (+) insulin stimulation, with standard deviations depicted by vertical bars. * Mean values were significantly different from those of the control group (two-sided Dunnett t test) (P < 0·05).
Total GLUT-4 mRNA and protein expression
The levels of GLUT-4 mRNA and protein were decreased in a dose-dependent manner (GLUT-4 mRNA, r − 0·899, P < 0·001; GLUT-4 protein, r − 0·964, P < 0·001) in skeletal muscle of rats; whereas levels of GLUT-4 mRNA expression increased in adipose tissue in the 10 % and 20 % (v/v) ethanol-loaded groups and GLUT-4 protein content was significantly increased in the excessive ethanol-loaded group (P < 0·05) (Fig. 6).

Fig. 6 Effects of ethanol on GLUT-4 mRNA expression (a) and protein content (b) in skeletal muscle and adipose tissue of rats. Muscle and adipose samples were collected. Real-time PCR and western blotting were performed to determine RNA and protein expression of GLUT-4. (see Materials and methods). C, control group; L, 10 % (v/v) ethanol-loaded group; M, 20 % (v/v) ethanol-loaded group; H, 33 % (v/v) ethanol-loaded group. Values are means with their standard errors depicted by vertical bars. * Mean values were significantly different from those of the control group (two-sided Dunnett t test) (P < 0·05).
Discussion
Published studies show conditional effects of ethanol on insulin sensitivity, determined by the factors of alcohol formulation, drinking patterns and the duration of consumption. In the study of Onishi et al. (Reference Onishi, Honda and Ogihara19), administration of 35 % ethanol in a liquid diet for 2 weeks effectively induced insulin resistance in Sprague–Dawley rats. Tomie Furuya et al. (Reference Tomie Furuya, Binsack and Onishi18) found increased insulin sensitivity in Wistar rats being treated with 3 % ethanol in drinking water for 4 weeks. The disparity in the results can be attributed to ethanol dose and the duration of ethanol administration. As the dose of ethanol may be a determinant factor of insulin sensitivity, intragastric injection was selected in the present study to minimize the potential impacts of other factors. Under the designed conditions, we found daily intragastric ethanol injection (10 %, 20 and 33 % v/v, 19 weeks) decreased insulin sensitivity in a dose-dependent manner (r − 0·842, P < 0·001) and 33 % (v/v) ethanol may cause necrotic/haemorrhagic injury in pancreas and significantly reduced islet β-cell mass.
Evidence from diabetic animal models supports reduced β-cell mass as a significant contributory factor to diminished insulin secretion in type 2 diabetes(Reference Wajchenberg26). In an in vitro study, ethanol was found to induce β-cell apoptosis in cultured insulinoma cells(Reference Dembele, Nguyen and Hernandez27). To our knowledge, the effect of ethanol on the β-cell mass in vivo has not been reported yet. In the present study we found that 33 % (v/v) intragastric ethanol-loading impaired the normal structure of pancreas and significantly reduced β-cell mass. Fasting plasma insulin level detected from the 10 and 20 % (v/v) groups were significantly higher than the control, while the level was relatively normal in the excessive ethanol group. One possible explanation is a two-stage response model of β-cells to different levels of ethanol. At the first stage, 10–20 % (v/v) ethanol initiated β-cell compensation by hypersecreting insulin to maintain normoglycaemia in the ethanol-induced insulin resistance state. At the second stage, 33 % (v/v) ethanol caused direct damage to islets and β-cells, and the reduced β-cell mass compromised the body's capacity to compensate for insulin resistance, leading to a fall of blood insulin level and a pathological higher level of fasting plasma glucose.
Skeletal muscle is responsible for at least 80 % of insulin-stimulated glucose uptake. Therefore, the insulin signalling and GLUT-4 content in skeletal muscle are determinant factors of whole-body glucose disposal(Reference Derave, Eijnde and Verbessem28, Reference Op 't Eijnde, Ursø and Richter29). The dose-dependent down-regulation of PI3K activity and GLUT-4 expression found in all ethanol-loaded rats in the present study indicated the reduced insulin-stimulated glucose uptake in muscle tissue. The present findings were partly coincident with Spolarics et al. (Reference Spolarics, Bagby and Pekala30) who found that alcohol administration (1800–1900 mg/l plasma for 3 h) attenuates insulin-mediated glucose use in skeletal muscle without a significant change of total GLUT-4 content in skeletal muscle. The disparity of the results is possibly due to the duration of ethanol; the duration of ethanol in the present study is longer (19 weeks). It has been reported that chronic alcohol intake (an alcohol-containing diet for 14 weeks) could inhibit muscle protein synthesis in rats(Reference Lang, Wu and Frost31).
In contrast to the changes in muscle, the down-regulation of PI3K activity and the expression levels of GLUT-4 mRNA and protein were not seen in adipose tissue. Conversely, the PI3K activity and the expression levels of GLUT-4 mRNA and protein were increased in the 20 % (v/v) ethanol-loaded group in adipose tissue. We presumed that the up-regulation of PI3K activity and the expression levels of GLUT-4 mRNA and protein in adipose tissue may be due to a tissue-specific response, which compensates for the decrease of whole-body insulin sensitivity in alcohol-loaded rats. The disparity of excessive alcohol on the PI3K pathway between muscle and adipose in the present study may possibly be due to the CAP/Cbl/TC10 pathway in adipose tissue. The CAP/Cbl/TC10 pathway is a second signalling pathway required for insulin-stimulated glucose transport(Reference Baumann, Ribon and Kanzaki32). Evidence indicated that skeletal muscle cells and adipocytes differ in their reliance on Cbl/TC10 in insulin-stimulated glucose uptake, and a TC10-dependent signalling pathway leading to GLUT-4 translocation in adipocytes may not operate in myocytes(Reference JeBailey, Rudich and Huang33). The present finding also partly confirms the observations of Becky, who found that chronic ethanol feeding in rats impairs insulin-stimulated glucose transport in isolated adipose tissue in a PI3K-independent manner and certificated the Cbl/TC10 pathway as a specific target of ethanol action(Reference Sebastian and Nagy34).
In conclusion, the present study indicated that chronic excessive ethanol intake is a risk factor for type 2 diabetes mellitus, and it is negatively associated with both insulin sensitivity and β-cell mass. The inhibitory effect of ethanol administration on insulin-induced PI3K activity and GLUT-4 expression in skeletal muscle could play a critical role in ethanol-induced whole-body insulin resistance in rats.
Acknowledgements
This work was supported by the National Natural Science Foundation of China (no. 30300282). There are no conflicts of interest. We thank Li Chen for reviewing the manuscript and providing helpful discussions. L.-N. Z. made major contributions to the experiment and the paper; X.-F. S., L.-P. H., X.-F. Y. and C.-J. Y. instructed on experiment design; and D. Y. helped on the animal model.










