Skeletal muscle is the major organ in vertebrates, especially in fish, which represent more than 40 % of the total body weight. It maintains protein metabolic homoeostasis of the whole body by acting as a major reservoir for amino acids and stores energy in the form of proteins to cope with nutrient deficiency(Reference Lecker and Goldberg1,Reference Wolfe2) . Branched-chain amino acids (BCAA) account for average thirty per cent of essential amino acids in skeletal muscle, and the breakdown of BCAA can produce alanine (Ala) which may be the most important source for gluconeogenic and protein synthesis substrate(Reference Nie, Ting and Wenju3,Reference Neinast, Murashige and Arany4) . BCAA are also important nutrition signalling molecules that have crucial regulating effects on protein synthesis, energy homoeostasis and nutrient-sensitive signalling pathways(Reference Jewell, Russell and Guan5–Reference Zhang, Lin and Hou7). BCAA are primarily catabolised and utilised as energy sources in skeletal muscle, on account of the expression of BCAA aminotransferase (BCAT2), which is a key enzyme that breaks down the first step of BCAA to produce glutamic acid (Glu) and branched-chain α-ketoacid, is high in skeletal muscle(Reference Harper, Miller and Block8,Reference Mann, Mora and Madu9) . In the fasting state, the transamination of BCAA is significantly increased in skeletal muscle, and then the metabolite Glu converted into Ala by alanine aminotransferase (ALT); eventually, the release of Ala is taken up by the liver as substrate for gluconeogenesis to maintain energy homoeostasis(Reference Harper, Miller and Block8,Reference Shimomura10,Reference Perry, Wang and Cline11) . The catabolic process of BCAA has been well known, but the adaptive regulatory mechanism and characteristic of transcriptional regulation during nutrient deficiency need further study, especially in aquatic animals.
Krüppel-like factor 15 (KLF15) plays a crucial function in regulating glycemic, lipid and amino acids metabolism(Reference Gray, Feinberg and Hull12–Reference Fan, Hsieh and Sweet14). Recently, KLF15 has been identified as a key transcriptional regulator in BCAA metabolism(Reference Fan, Hsieh and Sweet14). KLF15 can accelerate BCAA degradation and Ala production by upregulating transcriptional expression of Bcat2 in mice and rats(Reference Gray, Wang and Orihuela15–Reference Shimizu, Yoshikawa and Ito17). In addition, the mRNA expression of Bcat2 is significantly decreased in KLF15 mutant mice, and the ability to breakdown BCAA in muscle as substrate for gluconeogenesis is impaired(Reference Gray, Wang and Orihuela15). The catabolism of BCAA is enhanced, and the mRNA transcription level of KLF15 and Bcat2 is significantly increased in muscle of Oreochromis niloticus and Siniperca chuatsi after short-term fasting(Reference Li, An and Bao18,Reference Zhu, Hu and Zhang19) , indicating that KLF15 is involved in dynamic regulation of BCAA catabolism in fish in response to fasting. However, the molecular mechanism by which KLF15 is involved in this process is unclear.
Circadian rhythms, also known as the circadian clock, refer to changes in behaviour, physiology and molecules that occur on a cycle length of approximately 24 h(Reference Serin and Tek20). Many aspects of animal physiology and behaviour are coordinated with the light–dark cycle by circadian rhythm which is thought to be driven by molecular clock, that in mammals refer to the core clock genes(Reference Dibner, Schibler and Albrecht21–Reference Bumgarner and Nelson23). The circadian oscillator participates in regulation of energy homoeostasis by affecting food intake, expression and activity of hormones and metabolism-related enzymes(Reference Serin and Tek20,Reference Bray and Young24–Reference Schmutz, Albrecht and Ripperger26) . The peripheral circadian clocks play a unique and integral function in each of tissues and stimulate the rhythmic expression of specific genes participated in diverse physiological functions(Reference Richards and Gumz27,Reference Desmet, Thijs and Mas28) . Peripheral circadian clocks also have an important effect on the whole-body metabolism(Reference Bass and Takahashi29,Reference Asher and Schibler30) . More than 2300 genes have been identified as rhythmically expressed in skeletal muscle, and most of these genes have been identified as involved in metabolism, transcription and myogenesis(Reference Harfmann, Schroder and Esser22). When circadian clocks are disrupted, the type of muscle fibre, the structure of sarcomeric and the function of the muscle are all affected(Reference Andrews, Zhang and McCarthy31,Reference Dyar, Ciciliot and Wright32) . These data indicate a critical role for circadian clocks in skeletal muscle; however, further study is needed to reveal the regulatory mechanism of circadian clock in skeletal muscle.
The effects of fasting on expression of core circadian clocks and BCAA metabolism in skeletal muscle of fish have been studied(Reference Zhu, Hu and Zhang19,Reference Bao, Wang and Bin33,Reference Wu, Bao and Zhang34) ; however, the molecular mechanism of circadian rhythm regulating adaptive metabolism of fish skeletal muscle under fasting remains unclear. In this study, we investigated the expression characteristics of KLF15, Bcat2 and fifteen core circadian clock genes in fast muscle of Chinese perch (Siniperca chuatsi) during short-term fasting, then analysed the correlation between their expression to screen out the clock gene involved in dynamic regulation of BCAA metabolism, and finally demonstrated that the circadian rhythm regulates BCAA metabolism in Chinese perch during short-term fasting by Clock-KLF15-Bcat2 pathway.
Materials and methods
Fasting and daily rhythm experimental design and sample collection
The experimental work was performed following the guidelines approved by the Animal Care Committee of Hunan Agricultural University (approval number: 20190618). A total of 216 healthy juvenile Chinese perch with body weight of about 150 g were randomly divided into four groups (fifty-four individuals per each group) that fasted for 0, 1, 5 and 7 d, respectively. Among them, the 0-d fasting group was the normal feeding group without fasting treatment and was used as the control group. The fish in each group were kept in about 10 m3 tank which equipped with a flow-through water exchange and continuous aeration system. The fish were fed with live Carassius auratus twice a day at 08.00 and 17.00. Before the fasting experiment, the testing fish were acclimated to the above conditions for 1 month under 12.12 light–dark photoperiod, ZT0 was the time when light begins, and ZT12 was the time when darkness begins. After 1 month, tissue sampling was carried out at 0 (normal feeding), 1, 5 and 7 d of fasting. Before sampling, the fish were anaesthetised with 0·15 g/l tricaine methane sulphonate (MS-222). Fast muscle from dorsal myotomes were collected from five individuals in each group at ZT0, ZT3, ZT6, ZT9, ZT12, ZT15, ZT18, ZT21 and ZT24. All the muscle samples were snap-frozen in liquid N2 and then transferred to −80°C for preservation.
cDNA synthesis and quantitative real-time PCR analysis
Total RNA were isolated from Chinese perch fast muscle using RNAiso Plus (Takara) according to the manufacturer’s protocol. The RNA samples were quantified using a NanoPhotometer-NP80 (Implen), and equal amounts of RNA were reverse-transcribed using PrimeScript™ RT reagent Kit with gDNA Eraser (Takara).
Relative transcript levels were measured by quantitative PCR using SYBR Premix Ex TaqTM (TaKaRa). Primers for the qRT-PCR assays were designed using Primer Premier 5.0 software, and the sequences were shown in Table 1. Sequences of all genes used for quantitative expression analysis were referenced from the previous transcriptome database(Reference Wu, Li and Cheng35) and the Siniperca chuatsi genome database (http://genomes.igb-berlin.de/cgi-bin/hgGateway?db=sinChu7). The method of qRT-PCR was according to our previous report(Reference Harfmann, Schroder and Esser22). RPL13 gene was used as reference gene, and the relative expression level of target mRNA was calculated by R = 2−ΔΔCt .
Table 1. Primers for qRT-PCR

Branched-chain amino acids and BCAT2 content and enzyme activity determination
The BCAA contents in fast muscle of Chinese perch that fasted for 0, 1, 5 and 7 d were determined according to previous method(Reference Zhu, Hu and Zhang19). The sum of leucine, isoleucine and valine represents the content of the BCAA. The Bcat2 protein content in fast muscle was detected using a Fish Bcat2 ELISA Kit from Zhuo-Chai Biotechnology Institute, and the activities of ALT were estimated using commercial Alanine aminotransferase assay kit (Jian-Cheng Biotechnology Institute) according to instruction.
Dual-luciferase reporter gene assay
The in silico analysis identified two putative sites of Clock binding to KLF15 gene promoter and two putative sites of KLF15 binding to Bcat2 gene promoter. In order to determine whether Clock regulates transcription of KLF15 and KLF15 regulates transcription of Bcat2, the psiCHECK2-KLF15-WT, psiCHECK2-KLF15-Mut1, psiCHECK2-KLF15-Mut2, psiCHECK2-Bcat2-WT, psiCHECK2-Bcat2-Mut1 and psiCHECK2-Bcat2-Mut2 dual-luciferase reporter gene expression vectors were constructed. For overexpression of Clock and KLF15 in cultured cells, full-length Clock and KLF15 cDNA was subcloned into a pcDNA3.1-flag expression plasmid. As there is no muscle cell line of fish, the primary cultured Chinese perch muscle cells are unstable. So 293T cells, which are often used in luciferase reporter assays, were used for in vitro validation experiments in this study. Reporter plasmids were co-transfected into 293T cells with expression plasmids or control plasmids in 24-well plates using Lipofectamine 3000 (Invitrogen, L3000015). The luciferase activity was determined by Dual-Luciferase Reporter Assay System (Promega, E1910) according to the manufacturer’s protocol after transfection for 48 h.
KLF15 and clock gene knockdown
The knockdown (KD) of KLF15 and Clock genes in Chinese perch were achieved by injection of translation-blocking Vivo-morpholinos (Gene Tools) targeting the mRNA AUG translational start site or adjacent sequences. The antisense sequence used for KLF15 and Clock was 5′-TACCATCCCTGGATAGTGCCAAACG-3′ and 5′-CAGTGATTTGCTCTTTAGGCGTGA-3′, respectively. Healthy juvenile Chinese perch with body weight of about 150 g were chosen for Vivo-morpholinos injection experiment, and three individuals were injected in each group. Vivo-morpholinos were injected in dorsal muscle with 12·5 mg/kg (1·25 mmol/kg) body weight, and the control group was injected with equal amount of control oligos. Fast muscle samples were taken at 2 d after injection and stored at −80°C after snap-frozen in liquid N2.
Western blotting
For western blotting, proteins were extracted from fast muscle samples in RIPA lysis buffer, separated on 12 % SDS-PAGE gels, transferred to PVDF membranes by Trans-Blot Turbo and probed with primary antibody against KLF15 (Absin, abs113067, 1:1000), Clock (Proteintech, 18 094–1-AP, 1:1000) or β-actin (Proteintech, 20 536–1-AP, 1:2000). Samples were then stained with secondary antibody conjugated to HRP (Abbkine, A21020) at the dilution of 1:5000. The signal was scanned by the ChemiDoc XRS+ imaging system, and the grey values of protein signal were analysed by NIH-Image J software.
Statistical analysis
To detect variation in mRNA levels among different time points, statistical analyses were carried out with one-way ANOVA procedures by SPSS 19.0. The daily rhythmicity in relation to the expression of core circadian clock genes, KLF15 and Bcat2, was assayed with Matlab 7.0. To perform a cosinor analysis, the formula ƒ(t) = M + Acos (t/pi/12 –ϕ) was used, and the meanings of the letters in the formula refer to previous reports(Reference Wu, Bao and Zhang34). Significance of cosinor analysis was defined by the noise/signal of amplitude calculated from the ratio se(A)/A (hereafter referred as P-value). Expression was considered to display a daily rhythm if it had both P < 0·05 by ANOVA and P-value < 0·30 by cosinor analysis. The data were expressed as the mean ± se (n 5).
The data in gene knockdown and luciferase assays were expressed as the mean ± sd (n 3). Two-tailed Student’s t test was used for comparisons between two independent groups. For multiple comparisons, Duncan’s multiple range tests were used. P-values less than 0·05 were considered statistically significant.
The expression correlation between genes was tested by Pearson’s correlation test using GraphPad Prism 7.0 software. If the coefficient r is positive, it indicates a positive correlation between genes; conversely, the genes are negatively correlated. If 0·50 ≤ |r| < 0·70, there is moderate correlation between genes. If |r| ≥ 0·70, the genes were strongly correlated.
Results
Metabolic characteristics of branched-chain amino acids in fast muscle during short-term fasting
To analyse the metabolic characteristics of BCAA in Chinese perch during short-term fasting, the contents of BCAA, Ala and Bcat2, and the activities of ALT were measured in fast muscle. The BCAA content in fast muscle was significantly decreased after 1, 5 and 7 d fasting compared with 0-d fasting (normal feeding) (Fig. 1(a)). But in contrast, the Ala content in fast muscle was significantly increased after short-term fasting treatment (Fig. 1(a)). However, the content of Bcat2 protein was dynamically adjusted during fasting, with increased after 1 d fasting, reduced after 5 d fasting and then recovered to initial value (value in 0 d fasting) after fasting for 7 d (Fig. 1(c)). The activity of ALT showed increased after 1 d and 7 d fasting but unchanged after 5 d fasting (Fig. 1(d)). The results above indicate that the catabolism of BCAA in fast muscle is enhanced during short-term fasting, and the activity or content of BCAA metabolism-related enzymes has a dynamic adjustment process during the fasting state.
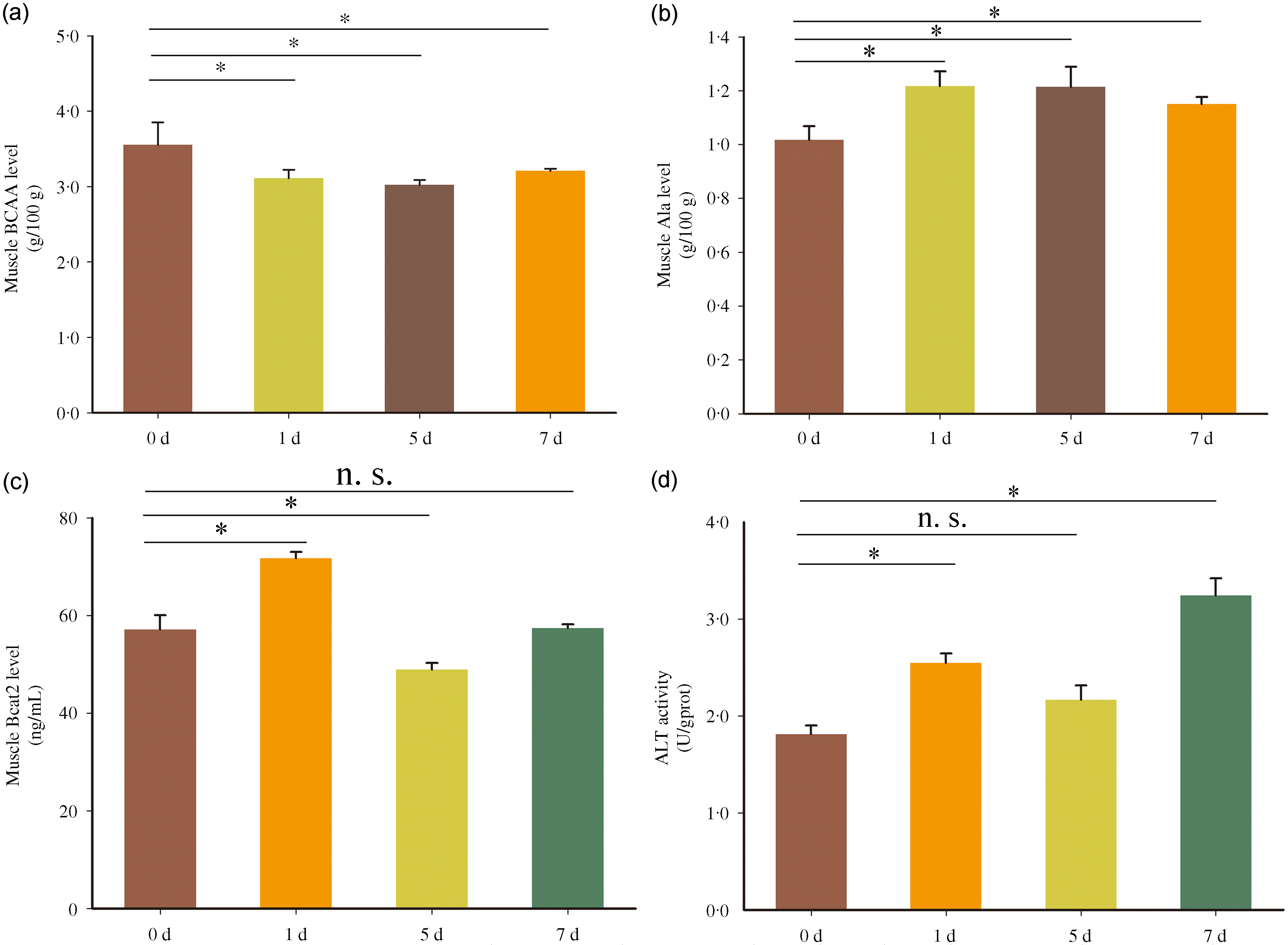
Fig. 1. Metabolic characteristics of BCAA in fast muscle of Chinese perch during short-term fasting. The A-D represents BCAA, Ala and Bcat2 contents, and ALT activity, respectively. Values in the figures are the mean ± se, n 5. The asterisk indicates a significant difference between two groups (P < 0·05). n.s. indicates no significant difference between two groups (P > 0·05). BCAA = branched-chain amino acids; Ala = alanine; ALT = alanine aminotransferase; d = days after fasting.
The rhythmic expression of Bcat2 in fast muscle of Chinese perch after short-term fasting treatment
To investigate whether BCAA metabolism is regulated by circadian rhythm, the expression of Bcat2 during a daily cycle was analysed in normal feeding (fasted for 0 d group) Chinese perch. The daily expression profile showed that Bcat2 was oscillating between day and night, and Bcat2 expression level was low during the day and high at night (Fig. 2). The result of cosinor analysis showed that the expression of Bcat2 displayed a significant daily rhythm (P-value < 0·30) with acrophase at night (ZT = 15·43) (Table 2). The result indicates that the BCAA metabolism in Chinese perch fast muscle may has a circadian rhythm. To further evaluate the effects of short-term fasting on rhythmic expression of Bcat2, the daily expression profile of Bcat2 was also analysed after 1, 5 and 7 d fasting (Fig. 2). After 1 and 5 d fasting, Bcat2 in fast muscle still displayed significant daily cyclic oscillations; however, the acrophases and amplitudes were changed (Table 2). The acrophase exhibited a left shift after 1 and 5 d fasting compared with normal feeding Chinese perch (Fig. 2 and Table 2). Whereas, fasting for 7 d disrupted circadian periodicity for Bcat2 which the rhythmic expression was disappeared (P-value = 0·63) (Fig. 2).

Fig. 2. Cosinor analyses of Bcat2 expression in fast muscle of Chinese perch during a daily cycle after short-term fasting. The values are mean ± se (n 5). Letters on the error line indicate significance markers, and different letters represents statistical difference between different time point (P < 0·05). The red dotted lines show the periodic sinusoids fitted based on the periodic parameters of each gene expression. ZT = zeitgeber time; 0, 1, 5 and 7 d = 0, 1, 5 and 7 d after fasting.
Table 2. Circadian parameters of Bcat2 and KLF15 expression during short-term fasting

The amplitude is half of the distance between two waveform peaks fitted. Median is the average of the curve. Acrophase is the radian corresponding to the time point of the highest amplitude. P-value is the noise/signal amplitude ratio in cosine analysis.
The rhythmic expression of KLF15 and correlation analysis of KLF15 and Bcat2 expression during short-term fasting
Considering that KLF15 can affect the expression of Bcat2, we speculated whether KLF15 is involved in regulating the rhythmic expression of Bcat2 in fast muscle of Chinese perch that under normal feeding or fasting state. To test this hypothesis, the daily expression profile of KLF15 and the cosinor analysis of its expression was analysed after 0, 1, 5 and 7 d fasting. The daily expression profile showed that KLF15 was oscillating between day and night, and its expression displayed a significant daily rhythm (P-value < 0·30) in fast muscle of normal feeding (fasted for 0 d group) and 1, 5 and 7 d fasting Chinese perch (Fig. 3). But the acrophases were changed during 1, 5 and 7 d fasting (Table 2). The results suggest that the transcription of KLF15 is also regulated by circadian rhythm, and its rhythmic expression can be dynamically adjusted with different periods of fasting. Next, the correlation between the circadian rhythmic expression of KLF15 and Bcat2 was analysed. In the normal feeding and fasting for 5 d fish, the transcript levels of KLF15 and Bcat2 displayed low positive correlation (r = 0·24 < 0·50). Interesting, KLF15 displayed moderate positive correlation with Bcat2 after 1 d fasting (0·50 < r = 0·57 < 0·70), and strong positive correlation with Bcat2 after 7 d fasting (r = 0·74 > 0·70).

Fig. 3. Cosinor analyses of KLF15 expression in fast muscle of Chinese perch during a daily cycle after short-term fasting. The values are mean ± se (n 5). Letters on the error line indicate significance markers, and different letter represents statistical difference between different time point (P < 0·05). The red dotted lines show the periodic sinusoids fitted based on the periodic parameters of each gene expression. ZT = zeitgeber time; 0, 1, 5 and 7 d = 0, 1, 5 and 7 d after fasting.
The expression of Bcat2 is regulated by KLF15
Although KLF15 has been reported to induce Bcat2 expression in mammals, it is not clear how KLF15 regulates the expression of Bcat2 in fish. To verify whether KLF15 regulates Bcat2 transcription, we analysed the 2-kb DNA sequence upstream of the Bcat2 transcriptional start site for evidence of G-rich element which has been reported as KLF15 binding site(Reference Du, Rosenfield and Qin36). The analysis identified two putative G-rich elements (designated as G-rich 1 and G-rich 2, respectively) in the Bcat2 gene promotor. The sequence of one site is 5′-GGGGAGGGGA-3′ (G-rich 1), and the other is 5′-AAACCCCCCCCC-3′ (G-rich 2, the complementary strand is 5′-GGGGGGGGGTTT-3′). Therefore, we used luciferase assays to determine whether these sites are involved in KLF15-regulating transcriptional expression of Bcat2. First, the dual-luciferase reporter vector containing G-rich 1 and G-rich 2 was generated, and the luciferase assays were performed in cells which transfected with the luciferase reporter vector and KLF15 overexpression vector or negative control vector. The result showed that overexpression KLF15 enhanced the luciferase activity of the reporter vector containing G-rich 1 and G-rich 2 element (Fig. 4(a)). Next, to confirm which element is regulated by KLF15, the G-rich 1 or G-rich 2 sequence in the reporter vector was disrupted, respectively, by site-directed mutagenesis. The result showed that G-rich 1 mutant abolished KLF15 regulation of the reporter activity, instead of the reporter construct containing G-rich 2 mutant (Fig. 4(a)), indicating that this G-rich 1 is a crucial site for KLF15-regulating Bcat2 transcriptional expression.

Fig. 4. The transcriptional expression of Bcat2 is regulated by KLF15. (A) Luciferase activity in cells transfected with KLF15 overexpression vector and reporter vector containing G-rich 1 and G-rich 2 element, or in cells transfected with KLF15 overexpression vector and reporter vector containing G-rich 1 or G-rich 2 element mutant (Mu1 or Mu2). (B) The KLF15 protein in control and KLF15 morpholino group by western blotting. (C) The relative protein level of KLF15 in control and morpholino group by grey scale analysis. (D) The mRNA expression of KLF15 and Bcat2 in control and KLF15 morpholino group. The asterisk indicates significant difference between two groups (P < 0·05). n.s. indicates no significant difference between two groups (P > 0·05). OE = over expression; Mu = mutant; MO = morpholino.
Considering that the 293T cells, which were used for in vitro validation in this study, differ significantly from fast muscle cells, a gene-specific antisense oligonucleotide, Vivo-morpholino, was designed to knock down endogenous KLF15 expression and to test the effect on Bcat2 expression. Compared with the control group, KLF15 protein expression level was significantly reduced in morpholino injection group (Fig. 4(b) and (c)), indicating the morpholino had successfully knock down KLF15. In addition, KLF15 knockdown significantly inhibited the transcriptional level of Bcat2 (Fig. 4(d)). Together, the in vitro and in vivo experiments demonstrate that Bcat2 expression may be directly regulated by KLF15. Interesting, the mRNA level of KLF15 was also decreased when knocked down KLF15 protein expression (Fig. 4(d)). Study in mouse has found that BCAA negatively regulated KLF15 expression(Reference Liu, Dong and Shao37); therefore, the decreased KLF15 might be attributed to the accumulation of BCAA when BCAA metabolism was inhibited by KLF15 knockdown.
The expression of core clock genes during short-term fasting and its correlation with KLF15
Previous study has identified that the KLF15 expression is regulated by core clock machinery in mammal(Reference Jeyaraj, Haldar and Wan38), and KLF15 is an important regulator of daily rhythmicity in skeletal muscle(Reference Tu and McKnight39). This suggests that the rhythmic expression of KLF15 is also controlled by core clock in Chinese perch. To identify which core clock gene regulates KLF15 expression, the rhythmic expression patterns of fifteen core clock genes were determined in fast muscle of normal feeding and short-term fasting Chinese perch. The circadian parameters of core clock genes in fast muscle of Chinese perch, which was fasted for 0, 1, 5 and 7 d, were shown in Supplementary Table S1–S4. The expression of tested genes showed a significant daily rhythm in normal feeding Chinese perch, except for Arntl2 and Cry-dash (Fig. 5 and online Supplementary Fig. S1). After 1 d fasting, the rhythmic expression of Cry1 was disappeared, and the expression of Arntl2 and Cry-dash began to show a rhythm (Fig. 5 and online Supplementary Fig. S2). After 5 d fasting, the rhythmic expression of Cry1, Rorα and Timeless (Tim) was disappeared (Fig. 5 and online Supplementary Fig. S3). The rhythmic expression of core clock gene was seriously disrupted during 7 d fasting, with only Clock, Arntl1, Arntl2, Rorα and Per2 still displayed significant daily cyclic oscillations (Fig. 5 and online Supplementary Fig. S4). These results indicate that the core clock genes are dynamic adjustment during short-term fasting. Among the fifteen core clock genes, only Clock, Arntl1 and Per2 had always displayed daily cyclic oscillations during the short-term fasting (Fig. 5).

Fig. 5. Cosinor analyses of Clock, Arntl1 and Per2 expression in fast muscle of Chinese perch during a daily cycle after short-term fasting. The values are mean ± se (n 5). Letters on the error line indicate significance markers, and different letter represents statistical difference between different time point (P < 0·05). The red dotted lines show the periodic sinusoids fitted based on the periodic parameters of each gene expression. ZT = zeitgeber time; 0, 1, 5 and 7 d = 0, 1, 5 and 7 d after fasting.
The correlation of circadian rhythm expression between KLF15 and 15 core clock genes was analysed. In the fast muscle of normal feeding fish, the transcript level of KLF15 displayed moderate positive correlation with Clock and Rorα (0·50 ≤ r < 0·70), and strong negative correlation with Timeless (|r| > 0·70) (Table 3). After 1 d fasting, Clock and Tim displayed strong positive and negative correlation with KLF15, respectively (Table 3). Meanwhile, Rorα, Nr1d2, Per1 and Per3 displayed moderate positive correlation with KLF15 (Table 3). After 5-d fasting, Clock displayed strong positive correlation with KLF15, Arntl1 and Npas2 displayed moderate positive correlation with KLF15, and Cry-dash displayed moderate negative correlation with KLF15 (Table 3). After 7-d fasting, the correlation between core clock genes and KLF15 was enhanced. Clock, Arntl1, Per2, Per3, Cry1 and Cry3 displayed strong positive correlation with KLF15, and the negative correlation between Time and KLF15 was disappeared (Table 3). The correlation analysis results showed that Clock always displayed positive correlation with KLF15, suggesting that the rhythmic expression of KLF15 is likely to be regulated by core clock gene Clock.
Table 3. The correlation analysis between core clock genes and KLF15 expression

The r values were set to define the degree of correlation, data are moderately correlated if 0·5 ≤ |r| < 0·7 and there is a strong correlation when |r| ≥ 0·7. If r is a positive number, it means a positive correlation, and the opposite means a negative correlation.
The expression of KLF15 is regulated by Clock
To verify whether the transcriptional expression of KLF15 is regulated by Clock, we analysed the 2-kb DNA sequence upstream of the KLF15 transcriptional start site for presence of E-box which is a canonical regulatory element for circadian clock. Two putative E-box elements (named E-box 1 and E-box 2) were identified in Chinese perch KLF15 gene. The sequence of one site is 5′-GCCACGTGCG-3′ (E-box 1), and the other is 5′-AACACGTGCA-3′ (E-box 2). Next, the luciferase reporter vector containing E-box 1 and E-box 2 was generated, and the luciferase assays were performed in cells which transfected with above reporter vector, and a Clock overexpression vector or negative control vector was performed. The result showed that overexpression of Clock increased the activity of luciferase in cells which transfected with the luciferase reporter construct (Fig. 6(a)). Then to confirm which site was the regulatory element recognised by Clock, the E-box 1 or E-box 2 sequence in the reporter vector was disrupted, respectively, by site-directed mutation technique. The luciferase assays showed that the E-box 1 mutant inhibited Clock regulated the luciferase activity, rather than the reporter vector with E-box 2 mutant (Fig. 6(a)). The results indicate that the E-box 1 is important for Clock regulation of KLF15 transcriptional expression. Furthermore, in order to evaluate whether Clock regulates KLF15 expression in vivo, a Clock-specific Vivo-morpholino was designed to knock down endogenous Clock expression. Compared with the control group, Clock morpholino injection significantly reduced the protein level of Clock and inhibited the transcriptional level of KLF15 and Bcat2 (Fig. 6(b)–(d)).

Fig. 6. The expression of KLF15 is regulated by Clock. (A) Luciferase activity in cells transfected with Clock overexpression vector and reporter vector containing E-box1 and E-box2 element, or in cells transfected with Clock overexpression vector and reporter vector with the E-box 1 or E-box 2 mutant (Mu1 or Mu2). (B) The protein expression of Clock in control and Clock morpholino group by western blotting. (C) The relative protein level of Clock in control and Clock morpholino group by grey scale analysis. (D) The mRNA expression of Clock, KLF15 and Bcat2 in control and Clock morpholino group. The asterisk indicates significant difference between two groups (P < 0·05). n.s. indicates no significant difference between two groups (P > 0·05). OE = over expression; Mu = mutant; MO = morpholino.
Discussion
The behaviour and physiology of organisms are affected by rotation of the earth(Reference Foster and Roenneberg40,Reference Jeyaraj, Scheer and Ripperger41) . In animals, diurnal cycle has driven the evolution of molecular clocks, syncing physiological and cellular processes to a cycle about 24 h. The past few decades have identified components and function of the central clock in mammals, but the function of clocks in the peripheral tissues is not fully understood, especially in non-mammal. As a highly adaptive and plasticity tissue, skeletal muscle has circadian rhythms(Reference Lazado, Kumaratunga and Nagasawa42). The daily rhythmicity of many clock genes was observed in skeletal muscle of fish(Reference Lazado, Kumaratunga and Nagasawa42–Reference Wu, Li and Cheng44), indicating that circadian rhythm has a potential role in regulating the physiology or metabolism in fish skeletal muscle. This study shows that circadian clock involves in regulating BCAA catabolism in fast muscle of Chinese perch during short-term fasting through a Clock crosstalk pathway to KLF15.
Precise regulation of metabolic processes is an important cornerstones of energy balance, and tight control of this homoeostatic process is essential for health and continuance of organisms. The circadian clock is the primary regulator of metabolism because there is growing evidence that the core clock machinery plays a central role in regulating metabolic homoeostasis(Reference Lamia, Storch and Weitz45–Reference Fan, Chen and Wang48). Previously, research in mouse and human has showed that nitrogen homoeostasis exhibits a 24-h periodicity and demonstrates that nitrogen homoeostasis is a conserved intrinsic circadian process in mammals(Reference Jeyaraj, Scheer and Ripperger41). BCAA are critical for the whole-body anabolism and energy homoeostasis, whether clock-driven oscillations in BCAA impact protein turnover is an attractive hypothesis. A recent study in muscular atrophy links disruption of circadian rhythm in regulation of skeletal muscle BCAA catabolism to severity of phenotypes(Reference Walter, Deguise and Meijboom49). Microarray analyses suggest that the BCAA α-ketoacid dehydrogenase is regulated by circadian clock at the transcriptional level(Reference Bailey, Udoh and Young50). Interesting, the daily expression profile of Bcat2 in Chinese perch fast muscle displayed significant daily cyclic oscillation, indicating that the BCAA metabolism in Chinese perch is under the control of circadian rhythm.
Previous studies have identified KLF15 as an important regulator of diurnal rhythmicity in skeletal muscle, heart and liver(Reference Gray, Wang and Orihuela15,Reference Tu and McKnight39,Reference Jeyaraj, Scheer and Ripperger41) . Research in mouse has identified KLF15 as a clock-driven peripheral clock factor critical for coordinating the transport of carbon skeletons and linked the clock to nitrogen homoeostasis by a KLF15-dependent way(Reference Jeyaraj, Scheer and Ripperger41). Furthermore, the rhythm of KLF15 is disrupted in some mouse lines with circadian clock gene mutant, which supports transcriptional expression of KLF15 is directly regulated by circadian clock(Reference Harfmann, Schroder and Esser22). This study showed that KLF15 regulated the transcriptional expression of Bcat2, and its expression exhibited 24 h periodicity and regulated by core clock gene Clock. Clock has been reported as a positive regulator of KLF15, and KLF15 rhythmicity is broken in core clock machinery mutant mouse(Reference Jeyaraj, Scheer and Ripperger41). The E-box is a critical cis-regulatory element that can be recognised by circadian clock to regulate transcription of flanking genes. The promoter region of mouse KLF15 gene revealed four canonical E-box regions for the core clock gene Clock(Reference Jeyaraj, Scheer and Ripperger41). Interestingly, two E-box binding sites were also found in 2 kb of the promoter region of Chinese perch KLF15 gene. Although only E-box 1 was verified to be the Clock regulatory site in Chinese perch, it suggests that the E-box is a conserved binding site for Clock regulation in at least vertebrates.
Fasting is a dynamic adaptive metabolic state when the intake of exogenous nutrient is lacking. In state of fasting, BCAA were preferential catabolism in the dorsal muscle of Carassius auratus gibelio as energy substrates, and BCAA catabolism in mice skeletal muscle is required to provide carbon substrates for gluconeogenesis to maintain glucose homoeostasis(Reference Gray, Wang and Orihuela15,Reference He, Li and Yan51) . Meanwhile, the circadian clock acts as an internal time-keeping mechanism to maintain homoeostasis in response to environment changings. Changes in expression of core clock genes in skeletal muscle after fasting have been reported in fish(Reference Bao, Wang and Bin33,Reference Wu, Bao and Zhang34) . This study also showed that fasting significantly changed the rhythmic expression of circadian genes, for example, the acrophases of Clock were dramatically changed during 1, 5 and 7 d fasting. This indicates that the circadian rhythm responds strongly to fasting and is highly dynamic and adaptive in response to different nutritional states. Although several reports have linked fasting to circadian rhythms(Reference Kawamoto, Noshiro and Furukawa52–Reference Longo and Panda54), it is unclear how fasting affects circadian rhythm to regulate energy metabolism homoeostasis. Recent report shows that fasting imposes specialised dynamics of transcriptional coordination between the circadian clock and nutrient-sensitive pathways, resulting in a switch to fasting-specific temporal gene regulation(Reference Kinouchi, Magnan and Ceglia55).
As an important regulator of cellular metabolism, KLF15 plays a crucial role in transmitting circadian rhythm to the release and utilisation of BCAA. Our study showed that the expression of KLF15 and Clock was highly responsive to fasting in fast muscle of Chinese perch. In addition, the expression correlation between KLF15 and Clock was enhanced after fasting treatment. The result indicates that fasting induces a switch in KLF15 expression through affecting the expression of Clock, thereby regulating BCAA metabolism. However, the fine regulatory network and mechanism of multiple circadian clock genes that coordinately regulate BCAA metabolism have not been revealed. For example, the Tim, a negative-feedback arm of the mammalian molecular clockwork, showed strong negative correlation with KLF15 during normal feeding, whereas the negative correlation was disappeared after 5 d fasting. This suggests that Tim may negatively regulate KLF15 under normal feeding condition and induce its negative regulation to be relieved under fasting condition, but the function and mechanism of Tim in regulating metabolic homoeostasis of BCAA need further study in the future.
Conclusions
In summary, the study showed that the transcriptional expression of core clock genes as well as KLF15 and Bcat2 was highly responsive to fasting, and the circadian clock involved in regulation of BCAA metabolism under fasting condition. Furthermore, we demonstrate that the transcriptional expression of Bcat2 is regulated by KLF15, and the transcriptional expression of KLF15 is regulated by Clock. Therefore, these findings suggest that fasting induces a switch in KLF15 expression through affecting the rhythmic expression of Clock, and KLF15 promotes the expression of Bcat2 to enhance the transamination of BCAA in fast muscle, then the Glu converts into Ala through ALT, finally the Ala releases into the circulation and absorbed by the liver as substrate for gluconeogenesis to provide energy for other tissues (Fig. 7). This study provides a mechanistic link between circadian rhythms and BCAA metabolism in teleost and opens a new field for the study in regulation of nutrient metabolism in fish.

Fig. 7. The mechanism of circadian rhythms regulates BCAA metabolism during short-term fasting. Glu = glutamate; ALT = alanine transaminase; Ala = alanine.
Acknowledgements
The authors would like to thank Dr Li Liu for her help in determination of amino acids contents. The authors are grateful to Dr Honghui Li and Dr Yajun Hu for their help in sample collection for the fasting treatment experiment.
This study was supported by the National Natural Science Foundation of China (No. 31972766; U21A20263; 31820103016; 32002370), the Natural Science Foundation of Hunan Province (2021JJ40629), the Scientific Research Foundation of Hunan Provincial Education Department (No. 20K014) and the fellowship of China Postdoctoral Science Foundation (2022T150207).
W. C., X. Z. and J. Z. conceived and designed the experiments. J. L., M. C. and Y. P. performed the experiments. J. L., X. Z. and L. B. analysed the data. X. Z. wrote the original manuscript. W. C., J. Z. and P. W. reviewed and edited the manuscript. All authors read and approved the final manuscript.
We declare that this study has no conflict of interest with other people or organisations.
Supplementary material
For supplementary material/s referred to in this article, please visit https://doi.org/10.1017/S0007114522003646













