The Chairman (Mr J. Taylor, F.F.A., President Elect): The topic for this evening is genomics and the potential impact on actuarial work. Concerns about some of the negative impacts led to the ABI announcing a moratorium in 2001. It said that anyone applying for life insurance need not reveal whether they had had a genetic test or, indeed, the results.
The impact of that moratorium was rather limited at the time, partly because the understanding of genetics and longevity was somewhat limited. The availability of genetic tests was somewhat limited. Fifteen years or more later, the impact on both information technology and biotechnology is profound.
The understanding now about the relationship between genetics and longevity is more advanced, largely due to the work of researchers like Peter Joshi and his colleagues. The availability of DNA tests has changed beyond recognition. There are television advertisements by companies like 23andMe, offering to do a health assessment based on your DNA for the order of hundred pounds.
It gives me great pleasure to welcome our opener, Angus Macdonald, from Heriot-Watt University. Angus graduated in mathematics from Glasgow University and then joined Scottish Amicable Life Assurance Society, qualifying as a Fellow of the Faculty of Actuaries in 1984.
In 1989, he moved to Heriot-Watt University, obtaining a PhD in 1995. He was appointed a professor only 5 years later, serving as head of department of actuarial mathematics and statistics from 2007 until 2013.
He was a member of the Faculty Council from 1998 to 2007. In 1999, he set up the Genetics and Insurance Research Centre, which has since produced most of the actuarial research on this subject, including two papers that won the David Garrick Halmstad prize in 2005.
Angus was elected a Fellow of the Royal Society of Edinburgh in 2006 and was awarded a Finlaison medal by the profession in 2011. He is currently editor-in-chief of the Annals of Actuarial Science.
Prof A. S. Macdonald, F.F.A. (introducing the paper): It is now about 25 years since genetic testing first came to the attention of actuaries in the mid-1990s. It was in a specific context at that time of mutations in single genes that were generally associated with severe disorder or extremely premature death. They had a clear pattern of Mendelian inheritance, which meant that the pattern of inheritance was evident in the families. These diseases had been studied for a long time. The only difference that the test made was that it turned a 50% chance of having an inherited mutation into certain knowledge one way or the other.
It led to immense controversy in the UK and in other countries. One of the most contentious points was insurers’ use of genetic tests. On the one hand, people feared genetic discrimination and loss of privacy. On the other hand, insurers feared adverse selection. This was dealt with in different ways in different countries. In the UK, it was eventually dealt with in a rather smart way by the Association of British Insurers. They obtained genetics advice from Professor Sandy Raeburn, who, sadly, died last year. They entered the moratorium, which John [Taylor] mentioned, in 1996. It has evolved since then, but it is with us still.
It has been successful in taking genetics away from the headlines in the UK. An important feature of the moratorium is that it was restricted to predictive tests only. It did not cover diagnostic and prognostic tests, tests which might in most circumstances be associated with a pre-existing condition. That has not always been the case.
The most recent example of government interest in the subject is in Canada, where Bill S-201 was passed in May 2017 and became the Genetic Non-Discrimination Act. That is completely sweeping. Insurers may not have access to any kind of genetic test results, predictive, prognostic or diagnostic, which, in my opinion, is a more serious issue than not having access to purely predictive genetic test results.
The question is, where has genetic epidemiology taken us in the past 25 years? It has taken us beyond these single gene disorders and into a considerable level of complexity called multifactorial disorders. Professor Joshi is going to tell us about that this evening.
It means that there are mutations in multiple genes, possibly quite large numbers of genes, each individually of small effect but in combination possibly of a considerable effect; and also interactions between genes and other genes, and between genes and the environment. So, you can see that this is a completely different world from the world of the single gene disorders.
It is also evident that we should not think of this multifactorial genetic world purely in terms of deleterious gene mutations. If we added up all the multifactorial genetics, then what we would get is the general population. They cannot all be bad. They must average out to some good and some bad. That also is a significant difference.
The complexity leads to some very difficult questions. There are questions of discovery, and questions of associations between genetic loci and an effect on what is called the phenotype, the physical organism.
What has been discovered is effects rather than causes, because, as Professor Joshi will explain, this kind of research is not identifying the genes responsible for a condition in the way that the single gene tests did.
There are quite often difficulties of replication. These are colossal experiments involving large numbers of subjects, and searching for relatively small effects. This is most definitely one of the branches of Big Science. It is not always the case that an effect that is detected by one study can be replicated in other studies.
Above all, there are difficult questions of interpretation of what a discovery of an association means in epidemiological terms and ultimately in clinical terms, and therefore in actuarial terms as well. The real difficulty, once the science has been done, is perhaps in the interpretation.
Generally, at a single locus, there will be two variants of a gene. They both lead to viable organisms or we would observe only one. Detecting these small effects is like looking for a very small signal among a lot of noise. It is very difficult to do in the first place.
The effect is expressed as a relative risk. There is a low-risk variant and a high-risk variant, and there is a relative risk of the high risk relative to the lower risk.
As difficult, if not more difficult, is the interpretation of the results at multiple locations where you believe that many variants collectively are having an effect on the phenotype, the condition or disease that you are studying.
How do they combine? It is generally assumed that relative risks can be combined multiplicatively. There is some evidence for that. When you think of the possibilities that arise from quite a modest number of genes, it is evident that the effects of combinations of gene variance must be based on a model rather than on observation.
What this leads to in combination, the adding-up of small effects, using the central limit theorem, is a normal distribution of risk where at the lower end you have people with lower risk in the population, at the higher end you have people with higher risk in the population, and in the middle you have population risk. When you aggregate all the risks, you must get the population that you observe.
As an example of the importance of the model in calculating these combined indices of genetic risk, which you then use as a relative risk in a model, I would refer to a study that I did with colleagues at Heriot-Watt University a few years ago of breast cancer and the effects of these multifactorial disorders.
At the time variants at seven different loci had been identified, associated with breast cancer risk. That did not come close to explaining all the inherited risks that were there to be explained. If you scaled up all the risks from the ones that were known, that suggested there were about 147 genetic loci which would account reasonably for the risk that had to be accounted for.
Three possible genotypes at each locus means the total possible number of genotypes in the model is 3 to the power of 147 and that is a very large number. On a conservative basis, it is larger than the number of people who have ever lived on the planet Earth by a factor of at least 1000 billion billion billion billion billion billion.
We are not going to say, “let us go out and get a sample of 100 people with this particular genotype”. The model is a central feature of the interpretation of the result of these multifactorial studies.
So, back to the question I started with: how is this going to translate into insurers’ risk and insurers’ use, or otherwise, of genetic test results? The context is important here. If you imagine a universe in which nothing but genetic information is relevant to the risk, and insurers are not allowed to use it, then, yes, that puts insurers in a very difficult position. But that is not the universe we are in, either 25 years ago or now.
Twenty-five years ago, we had a universe in which the available genetic information was single gene tests. These were not available to insurers, but everything else was – medical history, family history, and other things that insurers normally use in underwriting. Because of the rarity of these conditions, the impact was such that the industry could agree to the moratorium, and if there were any adverse selection costs, could absorb them. I have never heard of being noticed. The retention of the ability to use family history was far more important to the insurance industry at that time than being denied the use of genetic test results.
Now we are looking at a universe with genetic information at a much lower level of predictive power, if you like, being added to the mix of other things that insurers currently can use in their underwriting and reserving. If you think of a picture of the normal distribution of the risk, adding up large numbers of small relative risks, and multiplying them together, then, yes, there might be one or two people who have combinations of all the risky variants, a few dozen or so, but not many of them. Most of them will be somewhere in the middle, as in any Normal distribution. The kind of risks that that entails are of the magnitude that insurers have been used to coping with for as long as life insurance has existed.
I find the prospect of the new genetic information, whether or not it has been made available to underwriters, is unlikely to threaten seriously the viability of the life insurance industry. That was a conclusion that was true 25 years ago and although we now know a lot more, I think it has not changed.
I am looking forward to Peter [Joshi’s] comments on the subject and any light that he may throw on the risks that insurers may find before them, whether or not they know the genetic information that people themselves may know.
The Chairman: Peter Joshi started life as a commercial actuary reaching executive level at Standard Life and Tesco Bank, before returning to science and academia in 2011. Peter is now an Axa Research Fellow at Edinburgh University. His current principal research interest is the genomic basis for human lifespan, where an analysis of the survivorship of approximately one million UK Biobank parents is leading to fresh insights into how genetics influences longevity and how we might predict and perhaps shape healthy ageing.
Such is the interest in Peter’s work that each time he publishes new findings, it is not only covered in journals like Nature and on Radio 4 but also in the Sun and Metro! I am hoping that we will find out what the fuss is about.
Dr P. K. Joshi, F.F.A. (introducing the paper): Thank you for introducing the words “multifactorial disorders”. I want you to remember that multifactorial means more than one factor; in particular genes and the environment here. Also thank you for introducing the idea of lots of genes of small effect.
Ageing is better than the alternative. That is why I am so often exasperated by some actuaries and society in general with complaints about our ageing population. Increasing longevity is a sign of great progress in our society. At the same time, I do understand we are concerned about the pressures on our funding systems and perhaps the bigger question as to whether all the increase in lifespan that we are observing is an increase in healthy lifespan.
My research programme is about ageing. I am interested in measuring ageing, understanding ageing, forecasting ageing and, in due course, developing therapies that will allow us to influence ageing in a way that I hope will lead us to longer, more productive, healthier and, most of all, happier lives.
In recent years, my research has focused on genetics and genomics. Many of you are familiar with the idea that correlation is not equal to causation. Genetics, for a variety of technical reasons, is an exception to that. When I identify a genetic association between a region of DNA and lifespan, I am able more confidently to say that I have identified a more causal biological pathway than is the norm. That means that genetics is a particularly good instrument for understanding the causal pathways underpinning ageing.
So, what have I found? We are beginning to reveal how, and how much, DNA affects lifespan. I will show you how DNA can already be used to make meaningful life predictions at birth, at least at a group level, but at a level I think that would be of interest to actuaries.
Finally, I am going to look at the wider genomic revolution and talk to you about some of the real opportunities that that are presented to us but, at the same time, some deep ethical challenges and some of those are indeed actuarial.
So, what is deoxyribonucleic acid (DNA)? Most you will have heard that DNA is a double helix. The thing to realise is that the two helixes are mirror images of each other, that is mirror images in terms of the helix running around the outside, and mirror images in terms of the bonds between them which hold the two helixes together. That is true all the way along your DNA.
More importantly, it is these bonds, as I have called them, that are your DNA code. That DNA code is 3 billion bonds long, 3 billion letters long, 3 billion codons long, whatever term you want to use for it. You have two copies of that code – one you inherited from your mother and one that you inherited from your father. If you were to stretch the molecules end to end from one cell, it would be one and a half metres. There are 30–40 trillion cells in your body, all of which have copies of this molecule.
The key thing about DNA is its copying from cell to cell. It is that mirror image property that allows that copying to take place reliably.
It is a bit of a loose metaphor, but if you think of the easiest way to make a copy of yourself, it is to look in the mirror. That is part of the way that DNA manages to do copying.
So, if that is our DNA code, like any code the issue is how to decode it. Decoding part of a cell is called a ribosome. The DNA can be thought of as being fed into the ribosome in much the same way that tickertape might have been fed into an old-fashioned computer as computer code. The ribosome then translates that code into instructions to assemble amino acids. Those amino acids are then assembled into long chains in a specific order determined by the code.
Those proteins are what goes up to make your cells and tissues. Differences in those proteins are what goes to make up differences among us.
Does DNA determine lifespan? The historical starting point for this question is familial resemblance. In 1886, Francis Galton published a paper where he observed that very tall parents had taller children than average. He also observed that very tall parents had children who were not quite as tall as themselves on average. With some wonderful Victorian language, he talked about how height was regressing to the level of mediocrity. That is where the term “regression” came from. So, for anybody who has performed a linear regression, this is what was meant when it was first used.
The key point here is height runs in families; taller parents tend to have taller children, and you can do some sort of conventional regression.
The same is true of lifespan. Mothers who are living from 90 to 100 are having daughters who are living in the range of about 82 years; and, conversely, shorter-lived mothers having daughters who are living perhaps 75 years.
There is a big difference in the mother lifespan, but smaller differences in the expected lifespan of the children. This is the same process as Galton observed for height. That is true not just for mothers and daughters but for all the other relationships you might think of. It was true in the study that we did of 3,000 people in Fife, and the study of 2,000 people that we did in the Orkneys.
Lifespan runs in families, but how much? If you were lucky enough to have long-lived parents, say your mother lived 10 years longer than the average person in the population, by how much might you expect to live longer? Three quarters of a year. Not a great deal. Many people I talked to informally about this expected a bigger relationship. That is the general relationship across society that we would see. The same is true of fathers. If both parents had lived about 10 years longer than average, you might expect one and a half years increase for yourself.
Interestingly, and, for sceptics out there, importantly, we observe a similar correlation among spouses, and we are not expecting them to share an unusual amount of DNA. We do expect them to share environment. The question is: is the correlation between mother and daughter an environmental correlation or a DNA correlation?
Another way of addressing the question is a term that the geneticists call heritability. Heritability measures the proportion of the variation due to your genes. If the proportion is zero, I am saying that particular trait would all be about chance and environment. If, on the other hand, it was one, it would be a purely genetic trait. However, heritability varies by trait; and heritability estimation, for reasons I have already begun to allude to, is tricky and controversial. There has always been a great deal of controversy between social scientists and biological scientists about nature and nurture. That is at the heart of this estimate.
So, what do geneticists think about heritability of lifespan? Numbers range between 7% and 26%. More recent numbers have tended to be lower. If I were to come up with a single estimate, I would say about a twelfth, or about 8 per cent, of lifespan is genetically determined and the rest of it is determined by the environment. Chance itself plays a particularly large part.
How does heritability of lifespan compare with other things, such as height? Galton shows a good trait to start with. There is some uncertainty about the heritability of height. It is much more heritable than many other traits. You have probably seen that among people around you in your family.
Body mass index (BMI) is somewhat heritable; education level as well. We are confident that there is a genetic basis for that. It remains controversial whether that is 17% or 43% or somewhere in between. Then lifespan: we are confident that there is some basis in genetics, which I will illustrate more clearly in a moment.
So, does DNA determine lifespan? I have not mentioned DNA where I talked about heritability of lifespan. With our modern molecular techniques, what can we say about lifespan and DNA? Imagine your DNA code at one particular part of the 3 billion long letter code that went in to make you. What does it mean? I can tell you a few things looking at it, but I cannot tell you much about what it is doing. Some of my colleagues try to make codes like this into cells and understand what happens within the cells when that happens. Even then, if you understand what is going on at cellular level it is very difficult to take that up a level to a tissue, and so on, to an organismal level. And then again to a trait like lifespan. It is difficult to look at the code and to say what it does.
An alternative would be to ask what happens if we change one letter in the code. You might think of that in a computer program; what happens if you change one line of the computer program? What does that do? Run the computer program again and see what happens. Maybe run it thousands of times with and without the change and see what happens. That will tell you something about it.
You will be glad to hear that it is considered highly unethical to change one of your letters in your DNA and run you again thousands of times. That is not what we do as geneticists. We are in the fortunate position that nature provides us with a natural experiment of that form. Nature occasionally changes one of these letters. This is single nucleotide polymorphism (SNPs).
Occasionally across the genome, there is variation in the letters. So, this might have been your letters at this particular part of the genome. The person sitting beside you might have these distinct letters. If we were to analyse their traits versus your traits, we would not be able to come to that many conclusions. If we analyse enough people with enough precision, we can begin to understand what one of the single letters might be doing to that trait outcome.
The genome is the collection of your DNA. In the genome, there are chromosomes. The first chromosome is the biggest one. Each region of the genome has been associated with one particular disease, for example, digestive disease, cardiovascular disease, and so on. In the past 15 years, we have discovered thousands of robust associations between specific parts of DNA and these outcomes.
What do we know about lifespan? When I started my work in 2014, the cardiovascular disease was the only known robust association between lifespan and DNA. The reason for that contrast is partly the heritability; the genetic basis of lifespan is quite limited. There are so many other factors, and the small effects that Angus [Macdonald] talked about earlier.
Along the X-axis of Figure 1 is your genome. We see each of the different chromosomes represented along the X-axis. Every one of the spots is a region of the genome that has been tested for association with lifespan. Then on the Y-axis, the strength of the association is shown. There is one strong association, or one region of strong associations.

Figure 1. Genome-wide association meta-analysis of human longevity identifies a novel locus conferring survival beyond 90 years of age, Deelen et al. Human Molecular Genetics 23(16), 2014, 4420–4432, Figure 2, https://doi.org/10.1093/hmg/ddu139. © The Author 2014, used under the terms of the CC-BY 4.0 licence (http://creativecommons.org/licenses/by/4.0/).
As a genomicist, I consider the solid line as very important. Above that line we believe that we have a reasonably robust association, and below it we do not. We do not know whether the apparent association is just chance or it might be signal.
There is a suggestion of a signal, but it might just be noise and chance.
The people carrying out the study found 10,000 particularly long-lived people. They call those “cases”; “controlled” people had a normal lifespan. In both cases, they were still alive.
That worked to some extent. They found one genomic region and another robust association. That has been replicated.
However, it is very expensive to do. You must go out and find 10,000 people who are aged. You need to take their DNA, having obtained informed consent from them.
We have done something else. We have studied lifespan but because we recruit people when they are alive, we are going to have to wait a long time. So we ask them about how long their parents lived. We are able to make precise inferences from the recruit’s DNA of what their parents’ DNA is. If the parents were particularly long-lived and you have a certain kind of DNA, we can make some admittedly probabilistic inferences about what was going on with their DNA.
My PhD student, Paul Timmers, did most of the analysis of the data he gathered together of 1 million lifespans, which allowed us to do analysis of smaller sets with statistical robustness.
We found 12 robust associations between regions of DNA and lifespan. Typically, the effect of carrying one genetic theme relative to the other is about 0.3–0.5 years. A small effect, if you consider standard deviation of lifespan being, say, 10 years, and the total variation in lifespan would obviously be much greater than that.
The frequency of the beneficial variant and the deleterious variant are, broadly speaking, between 10% and 90%. Both are reasonably common. There are statistical reasons for that. I cannot find incredibly rare things that exist only once in a population. It is very difficult for me to do statistical inference on it. Similarly, that is the reason why I have not found smaller things. I need big enough effects even if they are small to find.
Finally, we had a look at the diseases. There is cardiometabolic disease as well as some others.
We then did something that is little complicated. Instead of looking at phenotypic correlations, we know the underlying genetics of lifespan and we know the underlying genetic basis of heart disease, so we can look at how the functions of those genes correlate. This genetic correlation has all the properties that you would expect of a correlation, but it only relates to genetic effect (Figure 2).
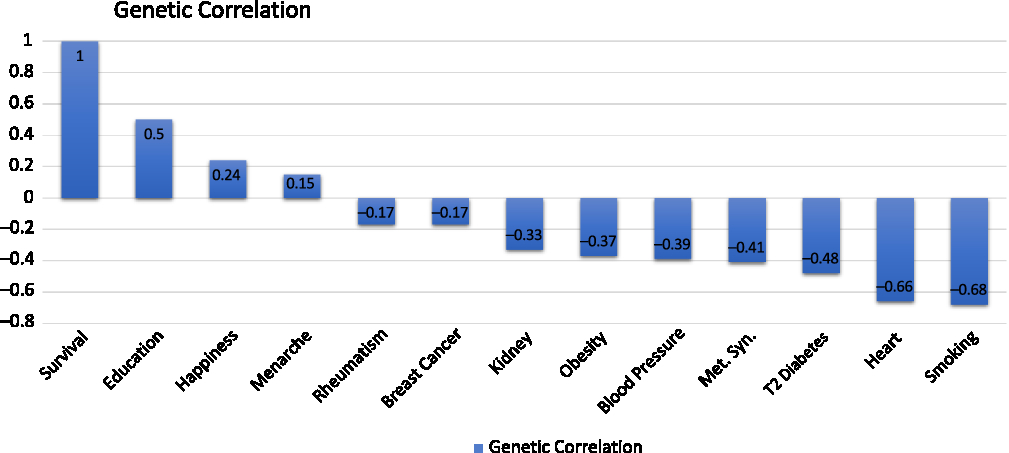
Figure 2. Genetic correlations (Reproduced with permission from Dr Peter Joshi).
The genetic correlations between survival, longevity, and various traits with heart disease, which shows a strong negative genetic correlation between susceptibility to heart disease and how long we live.
There is also a strong negative genetic correlation to propensity to smoke.
There is a positive genetic correlation between the genetic trait of the DNA influence on how much education you receive or how long you stay in education and survival. You may be even more surprised that there is a genetic trait of happiness. There are DNA variants that influence how happy you are. There is a genetic correlation between that and lifespan. There is a negative genetic correlation between educational attainment and happiness.
We have seen that major disease categories have been associated with genetic variation that we know affects lifespan: we then looked at disease categories and at the genetic variants that affect diseases to determine if they affect lifespan.
For Alzheimer’s disease, there is a particularly strong effect variant – the APOE variant – that is also protective of lifespan. The protection against Alzheimer’s disease by this variant is also protective against lifespan. We tested a lot of other variants and we saw what looks more like noise, certainly weak associations, between these other variants that affect Alzheimer’s disease, admittedly much less than the APOE variant and lifespan.
We then went on and looked at the genetic propensity to smoke and the associated suffering of lung cancer. Here we see what you might expect: a variety of genetic variants are known to affect smoking and lung cancer, in this case reducing them, increasing lifespan.
There are many genetic variants that we know influence susceptibility to cardiovascular disease. Each is reducing susceptibility to cardiovascular disease. Many cardiovascular disease variants affect longevity.
More surprisingly, we looked at a lot of genetic variants that are known to affect susceptibility to cancer. We have seen a much less clear line of sight between genetic susceptibility to cancer among these variants and how long we live. That is probably because the effect of the genetic variants in cancer may be smaller, and the effect of cancer on lifespan may be less than cardiovascular disease or at least it may act on only one pathway.
What we also see, which is quite interesting, is that the same gene here, a susceptibility to cancer, is associated with a longer life. That is happening because this gene is associated with a susceptibility to cancer for the protection against cardiovascular disease.
By getting the information across the whole genome, I can look at gene pathways and biological pathways. What we are seeing is the more significant associations are with lipid metabolism, with cardiovascular lipid, and with brain function. It looks like heart and brain genetics are influencing how long you live.
The effects vary by the stage of your life and by your sex. The Alzheimer’s variant affects women more and later in life. Conversely, the heart disease variant affects men more and earlier in life.
The evidence is DNA does influence lifespan. The next question, which is of interest to actuaries, is can it predict lifespan?
It is worth thinking about this framework that we have been in for 25 years or more: traditional genetic prediction. From an NHS website, which I have paraphrased, inheriting faulty BRCA1 genes raises genetic risk of cancer. Those genes may be passed to your children, and indeed you may have inherited them from your parents, if they carried them.
But it is far from the only cancer risk. There is a myriad of other genetic and environmental causes of cancer risk. The key point here is to speak to your general practitioner (GP) if cancer runs in your family. If you do not have cancer within your family to any great extent and you go to your GP and say “I want some genetic testing for cancer”, you will be turned away.
It is only if there is a significant amount of cancer running in your family that you would be sent to a genetic counsellor who would then look at the way it is arranged within your family. If it is mainly your partner’s family they are going to think that it is not relevant to you, and so on. But if it does run in your family, you may be recommended for a genetic test.
That test can be inconclusive. It can cause you anxiety to no purpose. It could lead to action. Angelina Jolie’s genetic counsellor obviously discussed action because she was found to carry the faulty BRCA1 gene and she had a mastectomy.
A genetic counsellor may offer you advice about telling your family and about family planning. There is a possibility, at least in the context of in vitro fertilization (IVF), of pre-implantation genetic diagnosis of the embryo, and only the risk-free embryos being intended to be implanted. That option has existed for a long time.
Ninety-nine per cent of the population might have the low-risk variant, which is why your GP might be unwilling to send you to see a genetic counsellor unless you have some evidence.
What has changed? I have shown you that a whole genome basis of understanding of the genetic basis of human lifespan has these particularly strong signals. Among the signals are noise and I have used some machine learning techniques to combine all of that and reading your DNA, and come up with a personalised score for you. That takes all that knowledge together. A personalised score will typically have a Gaussian distribution.
That has been done relatively recently in a paper that was published about 6 months ago for individual diseases. Some people did what we have described. They worked out a score – a polygenic risk score, because it dealt with many genes.
On the X-axis of Figure 3 was the percentile of the polygenic score for atrial fibrillation. That score could have been made at birth for these people. They just needed to take a cheek swab and run through the algorithm that I have described. No other information has been introduced into this, no information about socio-economic status, parent family history – just a cheek swab of DNA at birth.
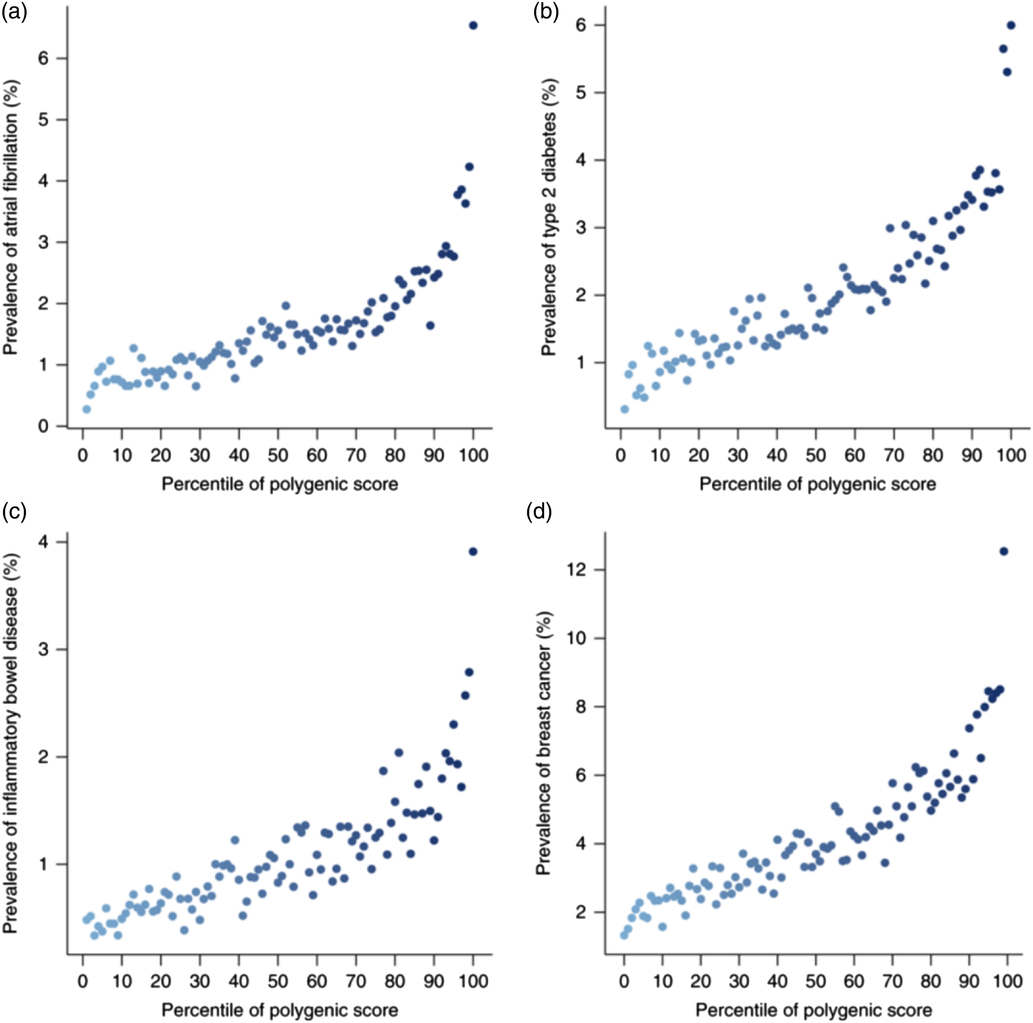
Figure 3. Polygenic risk scores (Reprinted by permission from Springer Nature: Nature Genetics, Genome-wide polygenic scores for common diseases identify individuals with risk equivalent to monogenic mutations, Khera et al., © 2018.).
What they found was that people who were in the safest few percentiles might have half a percent risk of atrial fibrillation, whereas people at the other end of the percentile spectrum would have a 4% risk, an overall eightfold increase in risk. Thinking about critical illness policies, that is clearly far from immaterial.
There were similar results for type 2 diabetes, for inflammatory bowel disease, and for breast cancer.
What about lifespan? We obtained polygenic scores just as I have described, using cheek swab at birth, or at any point in life. We can score people just basing their DNA into deciles.
What did we find in terms of survival differences? We found that the difference in survival time between the top and bottom decile for this score is about 5 years. That is not immaterial in actuarial terms. You are underwriting a set of annuities and there is a 5-year difference in expectation of life between one identifiable pool and another.
We saw that not just in the parents but in the subjects. We were able to do this better in Estonia because of the structure of their sample.
Focusing now on genomics more generally. Hardly a day goes by – certainly not a week – when there is not some announcement of some new genetic or genomic research and its potential impact on our society.
In 2001, the cost of sequencing your whole genome was $100 million. It is now $1000. Think about the progress that we have seen in electronic technology over that time. The effects of Moore’s Law for the genomic technology, has, albeit from a much higher base, arguably, proceeded much faster.
We are now in a situation where potentially every baby who is born in a “rich” hospital in America may see the genome sequence at birth.
The polygenic risk paper that looked at the relative risk of atrial fibrillation and breast cancer, and so on, is attracting a great deal of public interest.
You may have heard of a Chinese scientist editing the genome of someone. What I do has nothing to do with gene editing. I just measure your genes as they are. He edited the genes of an embryo and then implanted them. It is ethically controversial, because there was no real ethical consultation about carrying out this experiment in advance of it happening.
You will also have heard of gene therapy – the hope that we can inject different genetic material into ourselves, to compensate for a fault in our own genetic material or so that we might improve outcomes for ourselves.
The world is changing. In the past, genetic analysis was restricted to large effects within affected families. It was only possible for rare diseases and large effects like Huntington’s disease. That situation meant there was a successful ABI moratorium on insurers using genetic testing. The key to that was the fact that family history and other medical procedures were still disclosed to the insurers. We have had screening during IVF for rare large effect familial diseases.
Possibly, in future, everyone might be sequenced at birth. That would mean the day after your baby is born you would be able to calculate polygenic risk for educational attainment, longevity, risk of any given disease, and happiness. That process is also likely to throw up lots of variants of unknown significance.
You carry about 300 DNA differences that are not present in either of your parents. It is difficult to say what those DNA differences will be because they are unique to each person. Psychological impact of the scores of sector-related known diseases or those of unknown significance, the vast majority of which seem to have no effects, and the potential, depending on the way regulation develops, for an insurer/customer asymmetry of information.
I am not advocating these things. But this is a possible world in which we might end up and, unless society acts, without any choice.
It is possible, in principle, to screen for IVF any of these genetic risk scores. We have already seen gene editing happen. The controversial thing about that is the children that were born with these gene edits will pass those gene edits on down generations, and the potential for gene therapy for many major diseases, perhaps changing the outlook for gene longevity substantially.
I thank all the people and organisations involved in our research, in particular the Medical Research Council, the Axa Research Fund, and the University of Edinburgh for funding the research. Then the people and organisations who have provided data: the scientists that have gathered the data and those who have participated in the studies including UK Biobank, which has provided the bulk of the data that I have been able to analyse. Finally, I thank my colleagues at the University of Edinburgh and Lausanne.
In summary, we are beginning to understand direct lines of sight between individual DNA and longevity. The effects are small, though, at any particular site. However, combining all that information, we can now make, at least at a group level, meaningful distinctions in expectations of life and, in principle, those distinctions could be drawn up at birth.
Finally, the genomic revolution is presenting real opportunities and some deep ethical challenges to our society. Some of those are actuarial. I suggest that the technology is currently developing faster than the policy.
Mr A. J. G. Murray (opening the discussion): You talked about these genetic factors being quite small. I was wondering whether this is maybe just an effect of the fact that there are a lot of things that you can do to kill yourself and possibly if society was safer it may be more of an effect.
Dr Joshi: That is right. If we cite the susceptibility to death from buses, it would seem likely not to be at all genetic. We talk about the proportion that is due to genes and the environment. A very simple model you might think is that your lifespan depends on your genes plus environment plus chance, and the proportion would move towards genetics if we removed chance.
Or we could make our environment more homogeneous. If we all grew up in identical environments and had identical diets but different genetics, you would expect an increase.
Mr Murray: Would we expect something like that to happen?
Dr Joshi: I will leave each of you to form your own conclusions as to the future of our society and its homogenous heterogeneity. What we expect to happen is more and more therapies to come online. Whether or not that changes the balance I have just discussed, I am not sure.
The Chairman: I have heard you say before that if DNA at birth does say anything about the probability of age of death, it may be that some of those factors are already observable later in life, either through BMI or cardiovascular disease. What is the current thinking about how much additional information DNA gives beyond those observable facts later in life?
Dr Joshi: That is a good question and one to which at the moment there is no answer.
What John [Taylor] is saying is that if your genetic propensity to live long is about cardiovascular disease, we can measure your BMI at, say, 50. We can measure your cholesterol level. We can give you an ECG. Is genetics going to tell us anything more? Or has the genetics already had its effect and we are just waiting for that to emerge later?
That is a plausible model. It is certainly one of the next pieces of research that we plan to do. We have seen suggestions that in the context of cardiovascular disease, the clinical factors already explain a great deal of the variation, and genetic factors do not necessarily add that much more.
Having said that, as we get bigger and bigger studies, our understanding of genetics is getting better and better. We expect there to be a shift towards the explanatory power of genetics even in this context. How big that shift will be has yet to become clear.
Prof A. G. J. Cairns, F.F.A.: Does the Biobank dataset also include some socio-economic information? In your results, you had a 5-year range between the different deciles; if you also included some additional socio-economic information, would that allow you to widen the range and, if so, by how much?
Dr Joshi: Some members of this audience are probably more familiar with socio-economic underwriting and longevity than I am. Yes, the dataset does allow you to do that. Because we are focused on the DNA, we treat the socio-economic status as a noise variable in the analysis. So, we do include it. I have not separately analysed its explanatory power as opposed to DNA.
Mr M. C. Ledlie, F.F.A.: Do you have views on how insurers’ underwriting practices will or should change, and whether the ABI moratorium will, or should, be abandoned in the future?
Dr Joshi: This is a very difficult issue. There will be a great deal of hostility in society, and particularly among clinicians, to the use of genetic information in underwriting. I can see all sides of this.
Underwriters are used to bringing into account all sorts of factors such as socio-economic status. It is not easy to change socio-economic status, yet we are allowed to use that as an underwriting factor.
People see their DNA as particularly sacrosanct information. There is a lot of concern about personal privacy and DNA information, for example. There is potential customer/insurer asymmetry – information that the customer knows and the insurers do not.
At the same time, it is going to be seen by society as undesirable to use that information over which you have no control, and you are born with, in underwriting: that it is far from straightforward.
The results of our work, where we are seeing modest difference in long-term survival chances at the moment, may change and would suggest a storm brewing. I am not sure what the resolution is.
Mr P. D. Robertson, F.F.A.: Having had some involvement back in 1996, I maybe could make a comment and partly answer that.
In 1996, there were four clear groups: the insurers, otherwise known as the “bad guys”; there were the customers, otherwise known as the “poor unfortunates”; and there were clinicians, trying to help them. The argument was about single genes: if we cannot test these people, we cannot treat them, and there is no point in wasting money on treating people who do not have the condition.
Then there was the fourth group, the commentators, who were on the side of the clinicians and the poor unfortunates. The insurance industry was making its life difficult for no good reason.
Those four groups have now been joined by a fifth, and that is the people who produce the genetic tests. Then it was only the clinicians who were doing that. Now there is an industry designed to produce genetic tests, often producing information which is useless, like you are 3.6% North American Indian and 22.5% Pict, or whatever.
You have many genetic tests now that have nothing to do with treating people, but the commentators still think, as they did in 1996, that there is a killer gene for cancer; that there is a cancer gene out there, even though everything tells you there is not.
Until you can get over that, anyone who uses that information will always be the bad guy.
Dr Joshi: I agree with what you are saying, Peter [Robertson]. The other distinction, which is implicit in what you said, is in the family history question, as I highlighted earlier. Clinicians were only talking to people with a family history of these diseases. That is not the case now, or at least would not be the case in the scenario we are discussing.
The Chairman: We have some questions via social media. The first one is from Nicola Higgins who asks: “Are you aware of any studies on epigenetics and the impact on longevity? If so, what impact has been seen?”
Dr Joshi: With epigenetics, the DNA molecule is more complicated, inevitably. Along the outside of the backbone of molecules that give rise to what are called epigenetic effects – “epi” meaning above genetics. What those molecules do, among many other biological processes in our bodies, is switch on or off these genes.
Most of the epigenetic process is, for example, the one of how I decide – assuming I am a cell in your body – whether I am going to be an eye cell or a brain cell or a heart cell.
I have just said to you every cell in your body has the same DNA, but how does it decide what to be? It is epigenetic. It is because the cells code themselves with these epigenetic effects.
Epigenetics is fundamental to biology. For example, if you have a particularly poor diet and a high BMI, your epigenetics change. The way your cells behave changes.
Could epigenetic changes, and measuring epigenetics that differ across every cell in your body, in itself be prognostic of something? The answer to that is “yes”. If we measure epigenetics, we can construct something that is called a clock, and we can measure your age, because these effects change over your lifetime. It can infer biological age beyond chronological age, and that in turn is prognostic of lifespan.
Yes, epigenetic effects exist. There are bigger questions with much less consensus. There is much more coverage in the press about these effects than I think most geneticists are comfortable with. That is whether or not these epigenetic effects cascade down through generations.
The conventional wisdom, and the wisdom that I would run with, is, broadly speaking, epigenetic effects are not passed from generation to generation. So, we could not do a test at birth for these things.
As you age, as you consume a certain diet, as your cells differentiate, we get these epigenetic effects. They might be one way of measuring how old you are in the way that DNA does not. I could take your DNA, broadly speaking – it is not quite as simple as this – at birth and now and it is the same. However, epigenetics is evolving all the time within your body.
The Chairman: I have one more question from social media, from Steven Baxter. With dementia a growing cause of death, can Peter [Joshi] offer some thoughts on the likely genetic hereditability of dementia?
Dr Joshi: I cannot remember the heritability of Alzheimer’s disease. I suspect that it is in the range of about 30%, because most of these traits are in that sort of range. Part of that known heritability is it does not affect anybody who lives to age 70. In terms of outcome of lifespan, it affects a much smaller proportion of people in that range. As we live older and expect susceptibility to Alzheimer’s disease to play a much bigger part in how long we live, I am not sure if that, overall, means our genes will affect things more or less.
If you are suffering from less heart disease, which is heavily genetic, and more Alzheimer’s disease, which is also genetic, there is going to be a trade-off.
Dr Joshi: I have a question for you. Is a 5-year distinction in lifespan between the top and the bottom decile meaningful in underwriting terms either for annuities or for life insurance at earlier ages in terms of the implied hazard ratio, and also in terms of current underwriting practices?
Mr A. J. Rae, F.F.A.: I would say that it definitely is. The question is, if we were looking at pensioners or deferred pensioners, how much of that 5-year can we already infer?
If it were five new years of differentiation, it would be hugely valuable, let us say, for a pension scheme, if you are a bulk buyer for a billion pounds’ worth of pension liabilities, to be able to swab all the pensioners.
Prof Cairns: On the same point, the issue relates to the point at which people have the information about where within that spectrum they lie. At the moment, probably the vast majority of the population do not have that data. It is not immediately of consequence, but it will be in 10–20 years’ time when everybody gets this swab-at-birth prognosis.
If there are significant selection effects, at least we have the benefit of time to be able to think about that and to discuss what the consequences are. Now, there may be not such a big effect, but in the long run there could be.
Prof Macdonald: Following on from what Andrew [Cairns] said, in my earlier remarks, I imagined that universal genetic information is the only available information and there is nothing else, and that is exactly the scenario that Peter invented: the scenario of everybody having a swab, and having their genome sequenced, and then learning nothing else about people until they come to buy a pension at age 65. It is rather a case of what does genetics add to the conventional information available at the point of insurance purchasing process?
The Chairman: I should like now to introduce our closer, Alan Watson, who is a consulting actuary with Aon in Edinburgh. He advises clients on all aspects of pensions and has several scheme actuary appointments. He is also a member of the IFoA council management board and is leader of the Scottish Board.
Mr A. H. Watson, F.F.A. (closing the discussion): The discussion started with Angus Macdonald, who introduced us to genetics and actuarial work, and gave some of the background to insurers and genetic testing.
Angus reminded us that there are positives as well as negatives. He outlined many of the difficulties of using genetic information. He reminded us that other information was available and that was often much better than the genetic information.
Peter Joshi then took us through some of the research. He reminded us that increasing longevity is good news. We do worry that people living longer should not be seen as a bad thing. He took us through a high-level explanation of DNA and genetics. Few of us would doubt that lifespan would run in families. I think we were probably quite surprised that maybe only a twelfth of the influence is Peter’s opinion and there are many other more influencing factors. There are many small effects.
One of the disconnects is the DNA traits of happiness. We discovered the DNA traits of happiness. “How happy is the average actuary?” is something that he raised. Apparently, you are not happy if you are educated.
Combining these factors is important, rather than looking at individual bits of DNA. What is the combined information that we are getting from this?
The 5-year difference in life expectancy between the top and bottom decile sounds material. Alan Rae thought that it was material. But then we discovered later it was maybe not quite as much. We are near the point of genome sequencing at birth. That poses lots of questions. The challenge of technology ahead of policy is important.
Adam Murray made a valid observation about the other things that affect lifespan.
John Taylor, the chairman, raised the query of where pure lifespan and other influences are coming in. Where is the difference in pure lifespan influences and all these other influences? Peter Joshi has shown that they are all mixed together.
Andrew Cairns talked about the influences of socio-economic factors and Colin Ledlie asked the practical question about insurance underwriting.
Peter Robertson said that he was there in 1996. Educating those involved in the subject is going to be one of the challenges. There are many people who fear genetic information when it is probably not as fearful as we imagine. It is about educating people that this can be helpful.
We then had two questions from social media. Nicola Higgins asked about epigenetics. Peter gave us a thorough explanation of it. Then Steven Baxter came with the question about dementia, which is another challenge of the explanatory power of genetics.
Peter then posed the question: is the 5-year gap meaningful? Most people will agree that it is. Then it suddenly becomes more difficult. We accept that the challenge is to differentiate it from other factors.
As Angus Macdonald said at the end of the discussion, it is all very well to be swabbing everyone at birth but we are going to have all these other influences that are going to have equal or far more influence.
The Chairman: Thank you to all the presenters and all who contributed as well as the staff for organising the event.





