The invention of recombinant DNA techniques in the early 1970s consolidated the high-profile focus on molecular genetics, a trend under way since Watson and Crick's double-helical DNA model in 1953. With the tools of genetic engineering, biologists doing research on a wide variety of molecules (including enzymes, hormones, muscle proteins and RNAs, as well as chromosomal DNA), and any organism, could identify and copy the gene containing its ‘code’ and place that copy in a bacterial cell. At this point, the copied gene could be amplified, sequenced or analysed, or its product (usually a protein) expressed, purified and characterized. Initially, only a few molecular biologists and biochemists had the materials and know-how to do this. Many other life scientists sought this practical knowledge, to bring their labs into the vanguard of gene cloners. Manuals became a key part of this dissemination of expertise.
What did it mean to clone a gene? Simply put, cloning is copying. A gene would be isolated from all of the other genes in a cell, then inserted into a DNA ‘cloning vector’ that could replicate in a bacterial cell, so that the copied gene could be propagated indefinitely in a culture of the host cell, usually E. coli. In seeking to make copies of genes and move them around from organism to organism, biologists were inspired by bacteria, whose ability to exchange genetic material had been recognized in 1946.Footnote 1 It turned out that there were numerous genetic units (dubbed plasmids) that enabled gene exchange among bacteria in the real world.Footnote 2 By the 1960s, researchers were using these naturally occurring gene shuttles in microbes to identify, map and characterize bacterial genes.Footnote 3
Unsurprisingly, many biologists were more interested in studying genes found in humans and other ‘higher’ organisms (eukaryotes – plants, animals and fungi – as opposed to the one-celled prokaryotes, mostly bacteria). The discovery of bacterial restriction enzymes, which cleave DNA strands at specific base-pair combinations, inspired molecular biologists to attempt to use these as submicroscopic scissors.Footnote 4 In principle, if a researcher could identify and locate a particular eukaryotic gene, she could use a restriction enzyme to ‘cut’ it out of chromosomal DNA, insert it into a circular bacterial plasmid, then introduce the ‘recombinant’ plasmid into bacteria (see Figure 1).Footnote 5 That this could be done with genetic material from an animal was demonstrated first in 1973 by a collaborating group of scientists from the laboratories of Herbert Boyer (University of California, San Francisco – UCSF) and Stanley Cohen (Stanford). They inserted a frog ribosomal RNA gene into a customized bacterial plasmid. Not only was the inserted gene on its plasmid vector taken up and replicated by E. coli, but also the foreign DNA was transcribed into the corresponding rRNA product.Footnote 6
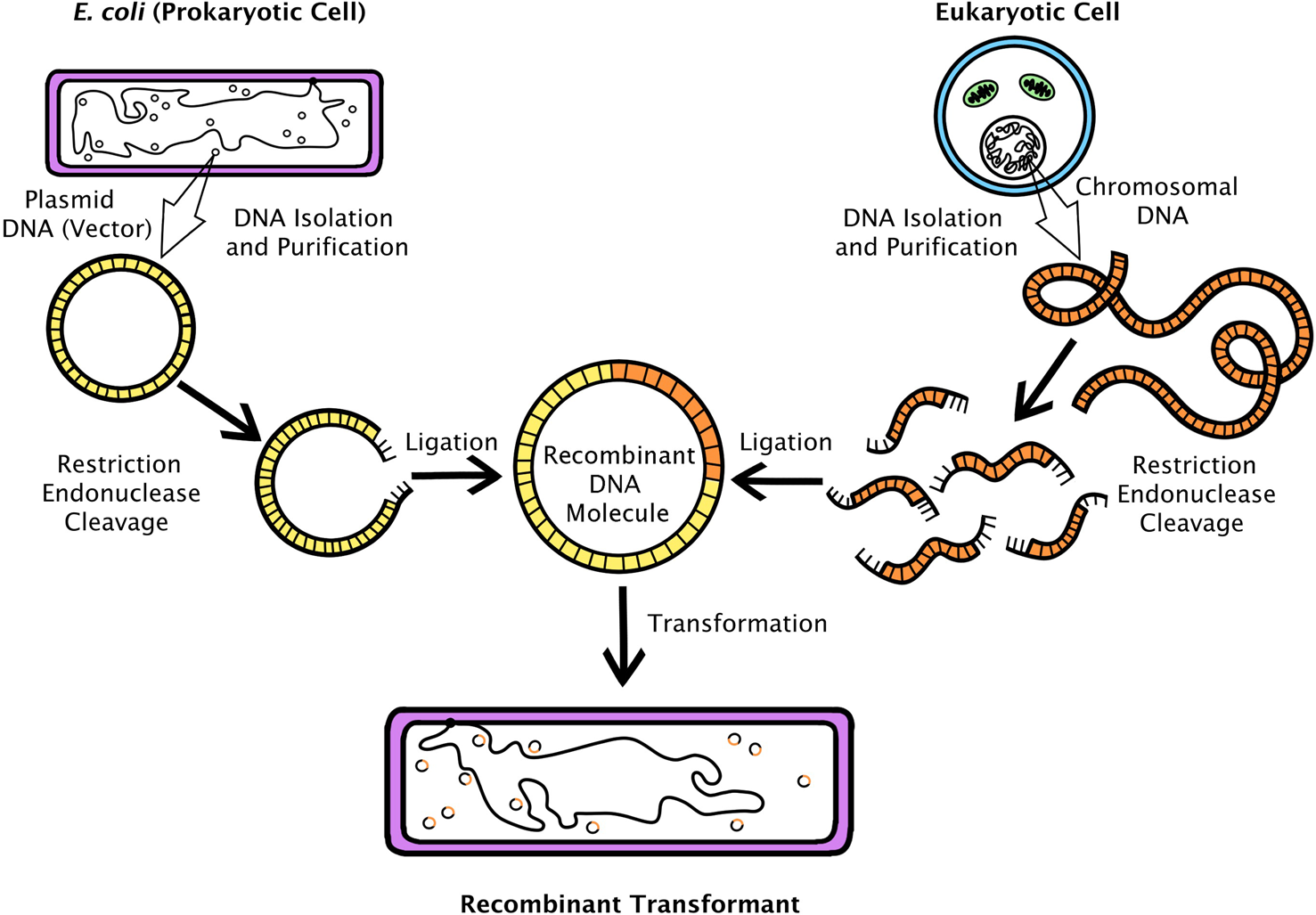
Figure 1. A typical recombinant DNA experiment depicting the cloning of eukaryotic genomic DNA fragments into a plasmid that is transformed into E. coli. Drawing by Georgia Creager.
However, cloning genes from higher organisms remained immensely challenging, for both intrinsic and extrinsic reasons. Intrinsically, it was technically difficult to locate specific genes in higher organisms. Both the human and mouse genomes, for instance, are 650 times larger than that of E. coli, so a given mammalian gene might comprise a ten-millionth of that organism's DNA.Footnote 7 The best way to identify the gene of interest was with a matching piece of nucleic acid, obtained by isolating the messenger RNA (mRNA) and using it to generate a DNA copy (cDNA). This was hard enough, but if a nucleic acid probe could not be produced, gene screening was even more arduous: every candidate clone had to be put into a protein expression vector, to search with antibodies or enzyme assay for the identifiable product.Footnote 8 Extrinsically, concerns about public-health hazards from genetically engineered pathogens led the National Institutes of Health (NIH) to require that recombinant work on higher organisms be done in biocontainment facilities with special ventilation systems and protective clothing, masks and gloves. Few such facilities existed in universities in the 1970s, as compared with military laboratories such as Fort Dietrich, set up for working with biological-warfare agents.Footnote 9 The impact on the field was dramatic, because the NIH funded nearly all of the leading academic molecular-biology laboratories in the US.Footnote 10
The first complete mammalian gene, that for mouse globin, was cloned in 1977 by scientists in Philip Leder's laboratory at NIH.Footnote 11 Shirley Tilghman, then a postdoc in the lab, recalls going with colleague David Tiemeier into a biocontainment facility with their cumbersome protective gear, and picking out tens of thousands of candidate clones, which appeared on Petri dishes of E. coli as plaques. Each plaque (a specially constructed lambda vector containing a random bit of mouse DNA) had to be transferred onto a nitrocellulose filter which would then be hybridized to a radioactively labelled globin cDNA probe.Footnote 12 They successfully identified the complete beta globin gene on a seven-thousand-base pair fragment of mouse DNA, but Tilghman remarked of the gruelling labour required, ‘it was ugly’.Footnote 13
Conditions for would-be genetic engineers improved in 1978, when NIH began relaxing the safety guidelines for recombinant DNA work on higher organisms. This meant that most work with recombinant DNA from higher organisms could be conducted in ordinary labs. Many more biologists wanted to learn these techniques, not only in academia but also in the burgeoning biotechnology industry.Footnote 14 One book became a canonical guide to this field, Molecular Cloning: A Laboratory Manual by Tom Maniatis, Ed Fritsch and Joe Sambrook, first published in 1982 (Figure 2). Including subsequent editions, this manual sold over 200,000 copies, making it a bestseller in the cottage industry of methods publications.Footnote 15 Drawing on both documentary sources and oral histories, this paper examines how protocols, recipes and pragmatic tips for gene cloning were shared, highlighting the role of published manuals as sources of practical knowledge.Footnote 16

Figure 2. Photograph of Tom Maniatis, Ed Fritsch and Joe Sambrook, Molecular Cloning: A Laboratory Manual, Cold Spring Harbor: Cold Spring Harbor Laboratory Press, 1982.
Learning how
By the late 1970s, scientists and journalists alike spoke of the ‘recombinant revolution’.Footnote 17 Most of the excitement revolved around the ability to clone and characterize individual genes from animals and plants, not least humans. The company Genentech, founded in 1976, led the race to clone genes and produce therapeutic proteins such as human insulin.Footnote 18 But the bounty was not restricted to the biotech industry. The techniques of genetic engineering had the potential to transform nearly every area of biomedical research. As one neuroscientist expressed it,
Imagine setting out to purify and characterize all of the proteins in a rat brain: each protein would require a specifically designed purification scheme, perhaps kilogram quantities of starting material, and the determination of each primary amino-acid sequence would be complex and time-consuming. To do this for each of the many thousands of proteins in a mammalian brain would be a daunting task indeed! Yet a few micrograms of rat brain mRNA contain molecules coding for every protein in the brain: by means of recombinant DNA techniques we can use the information encoded in each mRNA to investigate not only the structure of the corresponding protein but also its genetic regulation … This simple cloning procedure is the essence of recombinant DNA technology.Footnote 19
For biologists new to genetic engineering, assembling the needed materials required effort and expense. An array of specialized reagents, enzymes and plasmids began to be sold by companies such as New England Biolabs, which issued their first catalog in 1975. As mentioned earlier, finding a specific gene in chromosomal DNA was a major hurdle. The method used by Tilghman and colleagues to clone the first mouse gene effectively used a purification approach, fractionating the chromosomal DNA and then searching for the desired gene. Another approach was to construct a stable collection of DNA fragments from a particular organism, which could be screened for any individual gene. Such a collection was dubbed a genomic ‘library’. Tom Maniatis at Caltech, in collaboration with Arg Efstratiadis and others at Harvard, pioneered the methods for creating such a library and cloning genes from it.Footnote 20 He made his organism-specific libraries available to other researchers, even though each library was a biologically limited resource.Footnote 21 When Ed Fritsch, at that time a postdoc in Maniatis's lab, succeeded in developing a library of human DNA, it was reported in the Boston Globe, and researchers from many institutions began requesting it.Footnote 22
Even if one had all the materials at hand, however, reliable protocols and bench know-how could be just as challenging to procure. Even the cognoscenti struggled to keep up with new knowledge. As one of the cloners of the first mouse gene put it, ‘There was a lot of sharing “tricks” back then – it was pre-email of course. We all had folders or file boxes or notes stuck in notebooks of the various techniques that came as letters, phone messages, or notes from meeting talks.’Footnote 23 One key resource became available in 1979: volume 68 of the serial Methods in Enzymology, edited by Raymond Wu, was on Recombinant DNA.Footnote 24 Since 1955, Methods in Enzymology had provided standard protocols for biomedical researchers. This volume brought together the innovators of many of the key techniques for cloning genes. It provided a definitive set of methods, from their originators, for trained biochemists.
For DNA novices, courses were developed to teach recombinant techniques. In the autumn of 1979, Raymond L. Rodriguez, Robert C. Tait and other colleagues at the Department of Genetics at University of California, Davis offered a ten-week course entitled Advanced Molecular Genetics Laboratory. Rodriguez had been a pioneer in the field, having worked in Herbert Boyer's UCSF group that designed the most widely used plasmid cloning vector of that era.Footnote 25 His Davis course enrolled both undergraduate and graduate students, but there was, in addition, ‘a heavy demand for copies of laboratory handouts and protocols’. This inspired the publication of those in a course manual, Recombinant DNA Techniques: An Introduction, in 1983.Footnote 26 As it turned out, while the Davis course may have been the first to offer training in recombinant DNA techniques, its book was not the first entry onto the market.
Cold Spring Harbor Laboratory had been offering summer courses on new laboratory techniques since the 1940s. One popular course, Advanced Bacterial Genetics, already offered researchers a chance to learn how to identify, map and copy genes from microbes. These courses resulted in several important manuals for bacterial genetics, making the instruction available to many more scientists than could come to Long Island.Footnote 27 Since 1969, molecular genetic techniques for higher organisms were taught in an Animal (Tumor) Virus Course.Footnote 28 Nancy Hopkins co-taught the version of this course in 1979, but the enrolment was relatively low, in part because tumour virus know-how was available in ‘most academic centers’.Footnote 29 She argued that what was most needed at CSHL was a course on molecular cloning of eukaryotic genes, and that it had to involve Maniatis, whose lab was leading the development of recombinant DNA methods.Footnote 30 Maniatis, agreeing to teach it, recruited his postdoc Ed Fritsch to join as an instructor; Hopkins remained on board as the third member of the team that first year. Fritsch would put together all of the reagents and supplies for the course and adapted protocols from the Maniatis lab and the literature – no small task.Footnote 31 Helen Donis-Keller and Catherine O'Connell served as course assistants.Footnote 32
When CSHL advertised the postgraduate training course, Molecular Cloning of Eukaryotic Genes, 172 people applied for the sixteen spots. Thus even before it began, it immediately became the most popular course ever offered at CSHL.Footnote 33 Among the sixteen students selected for 1980, Robert Waterston would go on to a renowned career mapping the genome of a model worm (C. elegans) and, ultimately, playing a leadership role in the Human Genome Project.Footnote 34 Happily for the historian, he also kept his immaculately organized course notebook, as did Steve Goodbourn, now a renowned British virologist who took the course the second year.Footnote 35 Like other CSHL summer courses, the three-week schedule consisted of laboratory sessions and lectures by leading scientists in the field – in this case, twenty-one lectures (about half at 9 a.m., the others at 8 p.m.). Days were reserved for benchwork. Situated on a beautiful stretch on the coast of Long Island, Cold Spring Harbor Laboratory had a summer camp feel.
Both Hopkins and Donis-Keller recall that the night before the first class, just after they had finished setting up the teaching laboratory, there was a fire that filled the entire room with smoke, ignited by frayed wires in a piece of equipment.Footnote 36 They stayed late into the night cleaning up the mess. As Donis-Keller recalls of the rest of the course, while the lectures were great, it was a lot of trouble to get the experiments working, even with the expertise of the instructors.Footnote 37 The students were supposed to learn how to clone like the pros, construct a library in lambda phage, screen plaques with radioactively labelled nucleic acid probes and make a cDNA clone from messenger RNA, among other techniques. Likely reflecting the precedent of the tumour virology course, there were a number of lectures on that field's exemplars, such as SV40, adenovirus, RNA retroviruses and oncogenes. In contrast to these lectures on animal viruses, the laboratory exercises focused on cloning and manipulating eukaryotic DNA.Footnote 38
A student in the 1981 summer course (who had reapplied after not being selected in 1980), Gert-Jan van Ommen, remarked that socializing was a key part of what made the course so successful – amidst the long hours in the labs were breaks to swim in the Banbury pool, play volleyball games and attend barbeques (see Figure 3).Footnote 39 But it was the information conveyed in the course, going far beyond what was available in the published literature, that made the experience so valuable. Van Ommen recalls that ‘we were taught lots of tricks like cDNA synthesis, lambda cloning, making packaging mix, and cosmid cloning’.Footnote 40 Upon returning to the Netherlands, Van Ommen wrote up an extensive report ‘on all the techniques and tricks I learned there’, and sent it around the molecular-biology community in the Netherlands. ‘People were really over the moon with the report.’Footnote 41

Figure 3. Instructors and students working in the lab during the Molecular Cloning of Eukaryotic Genes course at Cold Spring Harbor Laboratory, summer 1981. The lack of lab coats reflects the casual atmosphere. There were more men than women students in the 1981 course, but almost equal numbers in 1980. Seated is Tom Maniatis; the man in the striped red shirt is Ed Fritsch. Photo courtesy of Gert-Jan van Ommen.
From course to book
Watson saw the opportunity to make this cloning know-how available to a wider base of users through publication.Footnote 42 Issuing an instructional guide on gene cloning from Cold Spring Harbor Laboratory would further consolidate the institution's reputation for being at the vanguard of molecular biology – and there was already a tradition from the Advanced Bacterial Genetics summer school of publishing course manuals as books.Footnote 43 Watson wanted Maniatis anchoring the team of authors, not only as an instructor but also on account of his renown in the world of cloning. But Maniatis had recently moved to Caltech, where he was busy with chairing an NIH study section, teaching and running his own lab. He only agreed to prepare a manual based on the course if he had significant help.Footnote 44 Watson persuaded Joe Sambrook, a talented and combative British tumour virologist at the lab, to join the effort. Although Sambrook never served as a formal instructor for the summer course, he was a leading research scientist at CSHL and a superb writer.Footnote 45 Fritsch, who was beginning a tenure-track faculty position at Michigan State University, remained centrally involved in the project.Footnote 46 The writing was already under way before Maniatis and Fritsch taught the course a second time with Doug Engels, in the summer of 1981.Footnote 47
None of the authors imagined the impact the ‘Maniatis manual’, as it came to be known, would have when it came out in 1982. For the second edition, Sambrook took a lead role and became first author, but the book's nickname stuck, irritating him.Footnote 48 Nancy Hopkins feels she should have been invited to participate as an author, having taught the 1980 course, though Fritsch and Maniatis are puzzled as to why she did not raise this at the time.Footnote 49 As the Preface makes clear, there were in fact a number of contributors who were not listed as authors. Maniatis, Fritsch and Sambrook thanked individual scientists (e.g. Brian Seed, Doug Melton) for providing particular protocols or for supplying the anthology of methods for a chapter (Nina Irwin).Footnote 50 In addition, Maniatis, Fritsch and Sambrook provided full references for methods adapted from the published literature.Footnote 51 Attribution was harder to ascertain for other protocols. As they stated,
We have tried to give credit at appropriate places in the text to the people who originally developed the procedures presented here, but in many cases tracing a particular method to its undisputed roots has proved to be impossible. We therefore wish to apologize – and to express gratitude – to those we have been unable to acknowledge for an idea, procedure, or recipe. Our major function has been to compile, to verify, and, we hope to clarify; less frequently we have introduced modifications, and only in rare instances have we devised new protocols.Footnote 52
In short, manuals raise the same problems of credit as other compilations, such as atlases and databases.Footnote 53 Though the authors (especially Maniatis and Fritsch) had personally developed key methods in the book, their role as authors of the manual made them stand-ins for many other scientists who had pioneered techniques. Not surprisingly, researchers began citing the manual rather than the original literature.Footnote 54 Sometimes, methods acquire an eponymous name, such as Maxam–Gilbert sequencing or Southern blotting. But for most methods, the successful circulation (and adaptation) of a lab recipe or method subverted conventional notions of authorship and credit.Footnote 55
That said, Molecular Cloning achieved a distinctive authorial voice, one aimed at the novice, but not the amateur. The three authors explain in the Preface that because ‘the manual was originally written to serve as a guide to those who had little experience in molecular cloning, it contains much basic material’.Footnote 56 Through chapter introductions that functioned in some respects like a textbook, the manual communicated enough about the science behind the recipes that users could troubleshoot the problems they encountered.Footnote 57 For instance, the first chapter on plasmids provided general information about these cloning vectors and genetic maps of the most commonly used ones, and outlined the three most popular methods of inserting a gene. As the authors note, ‘In principle, cloning in plasmid vectors is very straightforward’.Footnote 58 They then go on to enumerate the usual complications, and include a special section covering ‘Problems in cloning large DNA fragments in plasmids’.Footnote 59 This leads to sections on vectors used to handle larger pieces of DNA. A table lays out which of the four vectors was best for various experimental goals, such as cloning large DNA fragments, sequencing DNA or expressing foreign genes in E. coli.Footnote 60 This chapter repeats, and expands upon, material presented in the 1980 CSHL summer course.Footnote 61 Each of the other eleven chapters similarly instructs the reader on the relevant biology as well as giving practical advice in selecting methods. The text is also clear on omissions, such as ‘enzymes that find occasional use in molecular cloning but that are not necessary to carry out any of the procedures described in this manual’.Footnote 62 By contrast, chapters in the Methods in Enzymology volume 68 on Recombinant DNA, which also provided protocols, tended to be more like review articles in their comprehensive coverage. Readers might need to consult chapters by several different authors to understand the alternative strategies available for reaching their experimental goal.Footnote 63
The chapters of Molecular Cloning are organized around specific materials or procedures (e.g. Chapter 4, ‘Enzymes used in molecular cloning’; Chapter 7, ‘Synthesis and cloning of cDNA’). The overall sequence of information reflects the key choices involved in identifying and manipulating DNA, in the general order of steps required for cloning a gene. The second chapter, ‘Propagation and maintenance of bacterial strains and viruses’, provides basic instruction on isolating single colonies of bacteria and verifying strains through genetic markers, as might be taught in a microbiology course, as well as techniques more specific to molecular cloning, such as large-scale preparation of the vector bacteriophage lambda. This combination of instruction on basic lab methods with more specialized techniques distinguished Molecular Cloning from both textbooks and methods papers in the scientific literature.
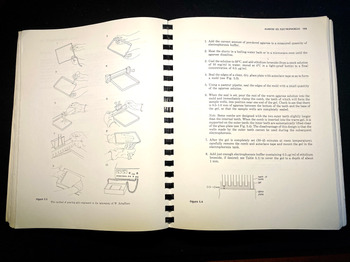
The book includes numerous drawings showing how to perform various procedures, such as recovering purified bacteriophage from a caesium chloride gradient, or how to pour an electrophoresis gel, in both cases showing where and how to position the hands (see Figure 4).Footnote 64 Integrated into the text are hundreds of tables and diagrams, including detailed genetic maps, as well as sample photographs of results. A well-furnished molecular biology lab of the time relied on a local machine shop to fabricate gel electrophoresis tanks; the manual included all the needed information in engineering drafting, with complete measurements.Footnote 65 Not all materials had to be custom-made, of course, and the manual mentioned many lab supplies by brand, such as Kodak X-Omat AR film, Whatman-52 filter paper, Sigma Type-III sodium salt DNA, and Dowex XG-8 mixed-bed resin. In addition, the authors included specific tips on how to keep costs down on restriction enzymes, the most expensive commercial reagents for gene cloning.Footnote 66
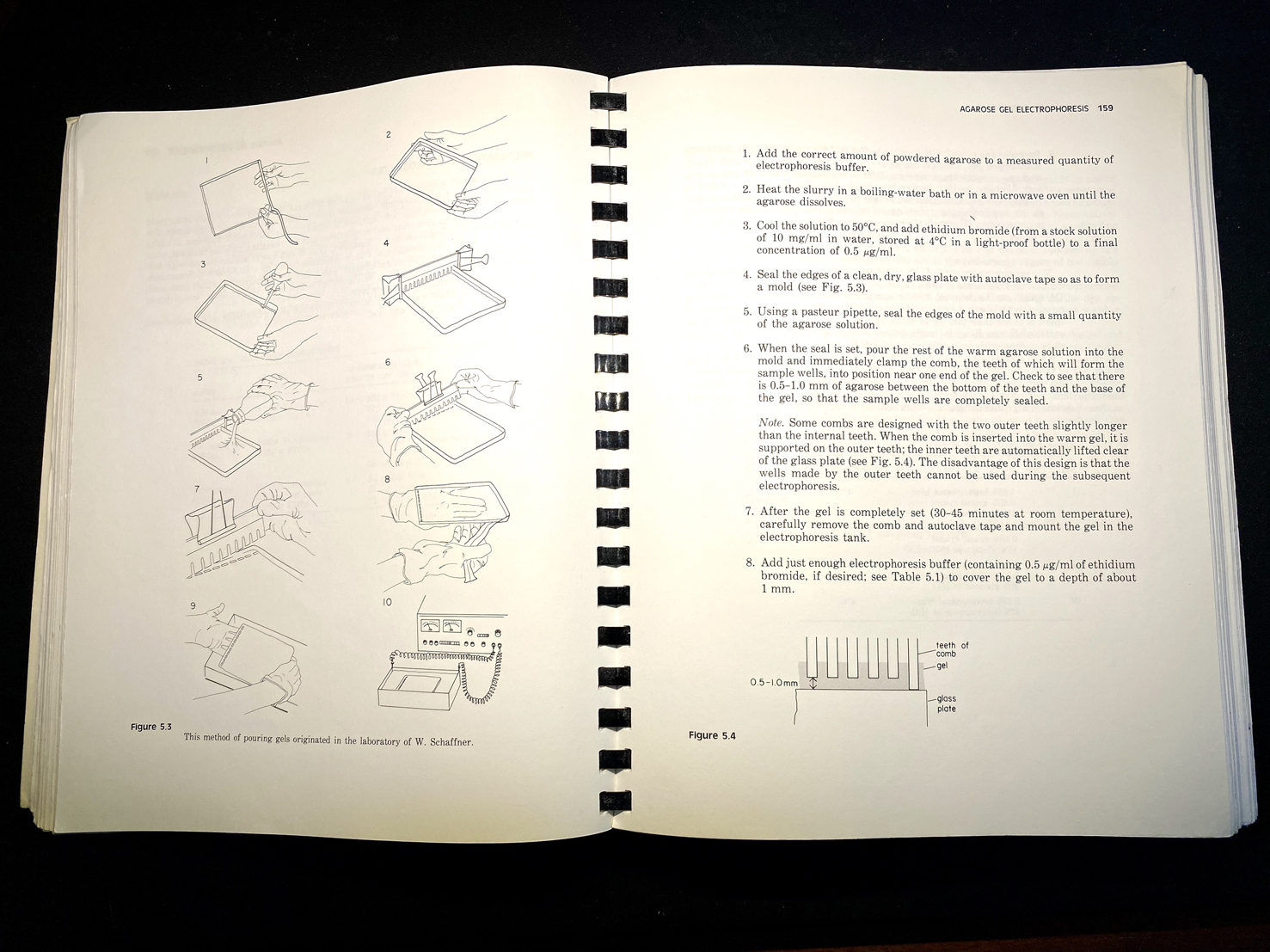
Figure 4. Instructions and diagram on how to pour an agarose gel for electrophoresis. In this set-up apparatus, agarose is used to separate a mixed population of nucleic acid fragments by length (in base pairs). Ethidium bromide is added to the gel to make the DNA visible under ultraviolet light. These descriptions include tips on the ‘combs’ that create the wells for DNA samples. Molecular Cloning, op. cit., pp. 158–9.
As a physical object, the 545-page manual brought together text, photographs, tables and diagrams, as well as countless technical symbols and some equations. The editing and layout of such a complicated publication must have been laborious, but the final product is clear and easy to read. Notably, the design is much more sophisticated and pleasing than that of the Advanced Bacterial Genetics manual that Cold Spring Harbor brought out just two years prior, which looked more like a print-out than a book, lacking any integration of images into the text.Footnote 67 Molecular Cloning did borrow one feature of that earlier manual, though: a plastic ring-comb binding (see Figures 2 and 4).Footnote 68 As David Crotty observed, some users complained that this binding served to ‘deliberately get the book to fall apart forcing you to buy a new copy’.Footnote 69 The advantage, however, was that the manual could be laid flat on a laboratory bench, a feature that the majority of users praised. The much-expanded second edition of Molecular Cloning appeared as three thick volumes in 1989; the plastic ring-comb binding remained.Footnote 70
Sales and rivals
Just as Watson expected, Molecular Cloning met a strong demand. There were orders for more than five thousand copies before the publication date.Footnote 71 Consequently, the press sold 5,113 copies the first month of its appearance, July 1982.Footnote 72 Sales remained robust that autumn, and in 1983 Cold Spring Harbor Laboratory Press sold more copies of Molecular Cloning than all of its other titles together (37,337 versus 24,234). The book went through four printings during its first six months, and three more were issued the following year. In explaining the surprisingly high sales number, the Annual Report 1982 points to the manual's ‘rapid adoption by many, many university courses’.Footnote 73
To be clear, students were not the only ones driving sales; working biologists were also buying the book in droves.Footnote 74 As a reviewer for the British Society for Developmental Biology put it, ‘no laboratory with any serious interest in molecular biology of development and their [sic] cloning should be without it’.Footnote 75 The testimony of historian of science Nick Hopwood confirms this view:
When I worked in developmental biology between 1986 and 1991 only two books had permanent places on my bench. The first, ‘Maniatis’, was a manual of the molecular cloning methods that our Cambridge laboratory was using to identify genes that specify muscle development in the South African clawed frog … I know the ring-bound recipes of Tom Maniatis et al. inside out.Footnote 76
The reliability of the protocols was a major reason for the book's popularity. As one admirer put it, ‘Just like the cookbooks of Betty Crocker and Fannie Farmer, the molecular cloning manual was chock full of recipes that worked’.Footnote 77 Another reviewer proclaimed, ‘In our laboratory, mirabile dictu, the procedures in this manual nearly always work’.Footnote 78
The trustworthiness of Molecular Cloning, however, did not solve the problem of keeping abreast of new and improved methods. Some researchers complained that the manual was out of date as soon as it was published.Footnote 79 Plans for a second edition, scheduled for 1984, were under way, but it was deferred for five more years as the first edition went through sixteen printings.Footnote 80 The rapid pace of change in molecular biology was blamed for the difficulty in updating the book. To take the most prominent example, the development of polymerase-chain reaction (PCR) by Kary Mullis and others at Cetus Corporation in 1985 made it possible to amplify a piece of DNA in a test tube, transforming cloning methods.Footnote 81 Thereafter, Crotty contended, a new Molecular Cloning manual without it would be ‘pointless’.Footnote 82
When the second edition of Molecular Cloning appeared in 1989, with the authors now listed as Sambrook, Fritsch and Maniatis, it was received just as enthusiastically as the first.Footnote 83 As a reviewer in Nature put it,
Few molecular biologists welcome publication of any of the many protocol books that promise to be the single source for their laboratory methods. For the most part, such laboratory methods fall far short of this goal. So why the excitement surrounding the long-awaited second edition of the classic guide, Molecular Cloning, which first appeared in 1982? The original version immediately filled the need for an anthology of laboratory procedures pertinent to the emerging field of recombinant DNA. With the 545-page spiral-bound paperback in hand, virtually any experimentalist could make a stab at cloning and have a reasonable expectation of success.Footnote 84
Both the first and second editions were reviewed in other languages, and translations appeared. A Russian translation of the first edition appeared in 1984; Chinese translations of the first edition came out in 1987 and of the second in 1992. Maniatis was told by travelling scientists in the early 1990s that ‘every lab they go to in China has the cloning manual on their desk’.Footnote 85
Reflecting its canonical status, Molecular Cloning was generally referred to by its users as the ‘bible’.Footnote 86 Extending this common metaphor, one biochemist made reference to ‘those who daily worship the Cold Spring Harbor idol’.Footnote 87 But the deity had rivals. Its main competitor in the first decade was Current Protocols in Molecular Biology, introduced in 1987 by a group of researchers based at Massachusetts General Hospital.Footnote 88 Sarah Greene was the original publisher, but the series was soon bought by Wiley. Rather than being written by three authors, this manual was produced by an entire team of scientists, who contributed individual pieces on various techniques. In addition, Current Protocols had a very different way of dealing with the rapid growth (and obsolescence) of techniques – the book was designed to be expanded via subscription. Through a quarterly update service, subscribers received supplements to insert into the original loose-leaf binder, which was separated into sections by preprinted dividers (see Figure 5). This meant that the table of contents also needed frequent updating. Five thick binders were published in the original series. The first volume covered most of the topics and methods in Molecular Cloning.

Figure 5. Volume 1 of Current Protocols in Molecular Biology opened to reveal how the protocols are organized by dividers in each three-ring binder. The topics in this volume are similar to those covered in Molecular Cloning. Frederick M. Ausubel, Roger Brent, Robert E. Kingston, David D. Moore, J.G. Seidman, John A. Smith and Kevin Struhl (eds.), Current Protocols in Molecular Biology, 5 vols., New York: John Wiley & Sons, 1987.
Maintaining the three-ring binders of Current Protocols as supplements arrived was inconvenient for users. Karen-Beth Scholthof, who was a postdoc in plant virology in Berkeley at the time, was responsible for updating the Current Protocols binders in the lab she worked in:
Your writing about Current Protocols reminded me of what a terrible system it was. I was assigned to add the quarterly updates (and remove the ‘old’ pages). This was such a hassle – having to take the entire binder apart to insert the new pages. The spiral-bound Maniatis was so much more user-friendly and the group annotations made it a kind of living document for the lab.Footnote 89
Given the unwieldiness of the Current Protocols loose-leaf format, in 1989 Wiley published Short Protocols in Molecular Biology: A Compendium of Methods from Current Protocols in Molecular Biology. This single-volume work was bound as a traditional text, with wide pages in a format that would prop open easily on the back of a lab bench. That same year the second edition of Molecular Cloning appeared, and more than seven thousand scientific articles cited the title that year alone. From 1991 to 2000, editions of Molecular Cloning were being cited over ten thousand times per year; by comparison, Current Protocols in Molecular Biology peaked at 2,520 citations in 1998.Footnote 90
There were many other manuals besides the rivals Molecular Cloning and Current Protocols – some geared towards students, with discrete lab exercises, and some to more advanced practitioners aiming to incorporate new techniques (e.g. PCR Cloning Protocols).Footnote 91 Others sought a place on the bench as essential reference tools. Bernard Perbal's A Practical Guide to Molecular Cloning combined up-to-date protocols with useful information on commercially available enzymes and cloning vectors.Footnote 92 In effect, this guide compiled what was available, but scattered, in free handbooks from companies such as Amersham, Boehringer and Pharmacia, as well as listing equipment needed for molecular biology and safety guidelines. Along similar lines, Terence A. Brown at the Department of Biomolecular Sciences at Manchester issued a book called Molecular Biology Labfax, a ‘compendium of essential information – on genotypes, reagents, enzymes, reaction conditions, cloning vectors, and suchlike – that is needed to plan and carry out molecular biology research’. As the author explained,
Some of this information is already available in cloning manuals, catalogues and possibly on pieces of paper kept somewhere safe, but tracking down exactly what you need to know takes time and can be a frustrating experience. With molecular biology becoming an interesting sophisticated science, an acute need has arisen for a databook to complement the traditional cloning manuals.Footnote 93
The notion of a ‘databook’ stands in an odd relationship to database, and the updating of such hard-copy publications attests to how slowly search engines came to replace other information-hunting methods.
The true test of any such reference work, however useful, was to become indispensable. As one reviewer remarked of Labfax,
To use books such as these to the best advantage, you need to spend a long time becoming familiar with them and, frankly, life is too short to master every book. I suspect that most mortals learn to feel their way around just one or two of these massive works and then use them over and over again: my favourites are Molecular Cloning: A Laboratory Manual and the 1989 Pharmacia Molecular Biology Catalogue (which, incidentally, is much better organized than the current catalogue!). Of course, Labfax contains a lot more facts than either of my two favourite works, but are they facts that I am ever going to want to know?Footnote 94
This comment crystallized the task facing publishers of compendia on cloning: how to determine exactly the current information most needed by biologists, in a format they would be happy to pay for, more than once as editions became obsolete?
The challenge of updating was more easily accommodated by the growth of multimedia technologies in the 1990s. By its third edition in 2001, Molecular Cloning: A Laboratory Manual came with a CD-ROM ‘Lab Book’. It also had an associated website, MolecularCloning.com, with the book's protocols and ‘discussion space for those asking questions or making alterations to techniques’.Footnote 95 However, the site did not have a ‘mechanism for continuously adding updated new material’, which was considered a major drawback given the technical feasibility of doing this online.Footnote 96 But who was to be the maintainer of up-to-date knowledge? The authors? The readers? In reality, there was not much community participation in the manual's online forum. Efforts by other publishers to create ‘yet another “myspace for biologists”’ also met with disappointment.Footnote 97
By contrast, ongoing maintenance of methods, recipes and protocols was vibrant at the level of individual laboratories. In fact, the more manuals and handbooks expanded to include more information, the greater was the need for abstracting the most essential instructions. In this sense authors of manuals face the same dilemma as those of handbooks, which tend to grow fatter with each edition. In response, CSHL Press issued a single volume called The Condensed Protocols from Molecular Cloning: A Laboratory Manual.Footnote 98 Some laboratories created their own solutions. As Karen-Beth Scholthof recounts about Molecular Cloning, ‘After it went to three volumes (Sambrook, although we continued to refer to it as Maniatis), we started formally writing and sharing lab recipes for common protocols. I also had an index card/recipe box with my favorite concoctions and cheat sheets for various calculations.’Footnote 99 At Washington University, Helen Donis-Keller developed a customized laboratory manual, specifically modelled on Molecular Cloning, with the methods and protocols her group relied upon.Footnote 100 As she commented,
We found that even though protocols were published by other labs or in the literature, we needed to test them ourselves and prepare updated standardized methods for use by everyone in the lab. The amounts of the various reagents we typically used might be different from the published protocols as well or we might have found that a different reagent than that specified might be more appropriate for our purposes. I really wanted everyone to work from the same set of protocols to maintain consistency and in order to make troubleshooting easier when things went wrong.Footnote 101
Her in-house manual, while ensuring uniformity in methods in a large (forty-person) lab, also made sure that know-how was not lost when people left. Each protocol was formatted in the same way, with the name of the person who wrote and tested it, the date, instructions, time needed, any special reagents or equipment required, and, at the end, the sources (which included, for example, Molecular Cloning).Footnote 102 As the annotations of ‘NOT DONE’ in the table of contents suggest, a customized manual of this sort was usable as a work-in-progress, updated and reimagined as needed.
The original challenge that had prompted Molecular Cloning, the need for instructions on how to copy genes from unsequenced chromosomal DNA, was simplified by the Human Genome Project, which was funded by the US Congress in 1988.Footnote 103 By the early 2000s, the full genomic sequences of several organisms were online and publicly available to researchers; biologists could retrieve their genes of interest more easily, and the comparisons available through sequence data inspired a new generation of in silico rather than in vitro experiments.Footnote 104 To be sure, Molecular Cloning expanded its reach for the age of genomics, and remains a relevant resource. But the massive shift of reference works to the Internet – including serials like Methods in Molecular Biology, started in 1983 – made the publication of manuals, and the release of new editions, less eventful.Footnote 105 In addition, the publication of protocols has also migrated back to journal publication, now also online. As Crotty notes, ‘The lessons learned from MolecularCloning.com led to CSH Protocols. We realized that if we were going to continuously add new material, we had to think of the project as a journal, and give it a full editorial staff’.Footnote 106 This new journal (and many other similar titles, such as Nature Methods) effectively moved the development and dissemination of molecular biology protocols back to the research literature, a full circle after the gene cloning manual's success.
Conclusions
Even among scientific methods, the history of gene cloning is unusual. This set of techniques went in the short space of ten years from being considered dangerous and in need of biocontainment to becoming adopted in laboratories throughout the world. There are many reasons for this radical and rapid shift, not least the politics of deregulation and the massive infusion of venture capital into biotechnology. But changing motivations for the spread of knowledge cannot explain how the practices spread so quickly, through a generation of technical workers, most of whom had completed their formal education. Manuals and other sources of how-to knowledge provided guidance to countless life scientists and technicians who mastered these cutting-edge techniques.
Among the manuals available, Molecular Cloning succeeded remarkably in becoming an indispensable element of the biology lab, often figured as the genetic ‘kitchen’. As one enthusiastic reviewer of the third edition, which was published in 2000, put it,
In every kitchen there is at least one indispensable cookbook. Sambrook and Russell's Molecular Cloning: A Laboratory Manual fills the same niche in the laboratory. Like its kitchen counterparts (e.g. Rombeck's Joy of Cooking) Sambrook's Molecular Cloning has information to help both the inexperienced and advanced user.Footnote 107
These comparisons are even more apt when one considers some of the supplies in molecular biology labs. Since the 1950s, bacterial geneticists had been using blenders to disrupt bacterial infection or mating. Methods developed in the 1970s for identifying genes using radioactively labelled nucleic acid probes employed ‘Seal-a-Meal’ bags for the hybridization step.Footnote 108 Carnation non-fat dry milk became the blocking agent of choice for Western blots.Footnote 109 There are countless other such mundane connections, though of course the traffic between lab chemistry and cooking is hardly new.Footnote 110
Yet seeing the manual as one's standby cookbook for gene cloning obscures a key difference, namely the rapid pace of change in DNA methods that molecular biologists faced in the 1980s. Of course, researchers had and have clever strategies for exploiting trustworthy protocols while also trying to master the latest methods. But there remains a central trade-off in laboratory how-to literature between reliability and state-of-the-art technique. During the height of the recombinant revolution, Molecular Cloning seemed able to provide both. But in biology, even the bible has to contend with obsolescence.
Acknowledgements
I thank Tom Maniatis, Ed Fritsch, Nancy Hopkins, Helen Donis-Keller, Gert-Jan van Ommen, Bob Waterston, Steve Goodbourn, Shirley Tilghman, Mike Levine, Karen Scholthof, Lynn Enquist and Arnold Levine for their generosity with time, information, materials and corrections; Kat Maxson Jones and Gina Surita for assistance with research and editing; and Jan Witkowski, Alex Gann, Marta Hanson, Mathias Grote and two referees for helpful suggestions. Gert-Jan von Ommen and Nancy Hopkins have allowed copies of their wonderful personal photographs from the 1980 and 1981 summer courses to be deposited in Cold Spring Harbor Laboratory Archives, along with the notebooks of Bob Waterston and Steve Goodbourn.