Impact statement
In fast-evolving research fields, involving software interoperability does not come first. Software developers are focused on developing the best algorithm possible. Researchers in the field have to choose which software to use and quite often they do not have the chance or the skills to switch to another without starting over. In case they make the switch, they are faced with serious issues like traceability, data, and metadata conversion which will complicate their research and divert them from their main activity. Scipion3 fills this gap bringing interoperability among software, traceability, reproducibility, and versatility.
1. Introduction
Cryo-electron microscopy (cryo-EM) has become a mature structural biology technique to solve high-resolution biological structures(Reference Namba and Makino1). Briefly, four consecutive tasks complete the cryo-EM pipeline: sample preparation, image acquisition, image processing, and structure modeling(Reference D’Imprima and Kühlbrandt2–Reference Sorzano and Carazo4). The last two steps fall into the image or data processing activity. Over the last decade, many algorithms have been published to assist structural biologists in achieving protein structures. However, the nature of these image-processing programs differs in many aspects: in terms of execution, some can be called from the command line in an automated way while others need graphical user interaction, and some require large computational clusters and long execution times while others are swift. Concerning imaging conventions, each program assumes a specific location of the origin of the images, contrast, preprocessing steps, the geometrical meaning of shared parameters, and exclusive parameters uncommon to other programs. Regarding software, programs depend on different programming languages and libraries, which in turn may conflict with other versions of the same library required by other software; and so forth. All these differences hinder interoperability among the various programs. Each one of these programs has different strengths and weaknesses. Robust image-processing pipelines need to combine various software packages, compare their results as a way to validate the outputs and continue only with those parameters that have been consistently found by two or more different algorithms(Reference Sorzano, Jiménez-Moreno and Maluenda5).
An image-processing workflow engine aims to lower the interaction barriers among the different software packages. With this aim, Scipion wraps the image-processing steps into small modules called protocols. Most protocols work similarly: (Step 1) Converts the input to the file format and imaging conventions assumed by the image-processing program. (Step 2) Sets up the execution environment needed by the program and invokes it with compatible input data and metadata. (Step 3) Collects the output from the program and keeps annotations about the output data and metadata, possibly converting part of the output metadata into a common representation that can be subsequently adapted to the input of the following protocol. The internal representation of the different objects within the workflow engine must be flexible to keep (a) the shared information in a standard representation that can be converted to any of the integrated programs and (b) the uniquely defined information that is only used by a specific software package that may be needed in subsequent steps. This flexibility significantly restricts how the workflow engine can internally represent the information (see Figure 1).
In addition to the interoperability requirement and the need to construct robust image-processing pipelines, there are other needs that any image-processing workflow engine must fulfill (in the following list, we will include interoperability for further reference):
-
A. Interoperability. Guarantee that the output of one of the image-processing steps can be the input of the next one. Interoperability involves file format and geometrical conventions, as well as preprocessing conversions. It may also include providing an environment compatible with the different software packages (directory structure, filename conventions, system environment, etc.).
-
B. Smart object model. Different entities along the path have different meanings and can be used for various purposes. These entities may follow an ontological hierarchy. For instance, we may have a generic “set of images” that is further specialized into “set of movies,” “set of micrographs,” and “set of particles.” Despite all of them being sets of images, their metadata differs among them.
Tasks may be applied to some of the object types but not to others. For example, when a protocol defines its inputs, it may also limit its input types. Additionally, there may not be a one-to-one relationship between an object and a file. For instance, a set of images may involve a single file (with the entire stack of images), various files (each image stored independently), or even a set of files and a set of associated metadata, which, in its turn, may be kept in one or various files, or even as a set of entries in a database. The workflow engine must be capable of supporting several formats, as each software package may make assumptions about the physical organization of the data.
-
C. Gluing. Along with the execution of a complex image-processing workflow, housekeeping and organizational functions are required. For instance, applying a constraint to a set of images. These constraints are generally applied to the metadata associated with the images such as alignment or acquisition parameters, classification into different subgroups, quality factors, and so forth. Since these operations are not usually provided by the underlying software packages, primarily focused on scientific calculations, the workflow engine should implement them to facilitate flexible image processing and rich analysis capabilities.
-
D. Comparison. Since in many cases, the same task can be solved by several algorithms, a crucial family of operations that the workflow engine can provide is comparing the results of those algorithms to identify metadata incorrectly estimated.
-
E. Wide set of domain-specific operations. The availability of the required tools for image-processing analysis is partly behind the success of a workflow engine. In cryo-EM image processing, Scipion integrates over 60 software packages and over 1,000 protocols(6), all related to the computational analysis of macromolecular structures. This richness allows very sophisticated image analysis pipelines thanks to the possibility of combining multiple software packages along the path and comparing their respective outputs.
-
F. Execution in streaming. The acquisition of a cryo-EM dataset typically takes a few days during which some TBs of data could be generated. To identify problems related to the acquisition or the sample quality early and take corrective actions, it is essential to start processing the data as it is produced. Nevertheless, this poses a challenge to the design of the workflow engine as the entities flowing through the pipeline are not “closed.” Instead, they are continuously updated with new data that need further processing. This fundamental characteristic is specific to image processing and is missing in most alternative workflow engines, which typically assume that the input datasets are static.
-
G. Smooth integration in high-performance computing (HPC) environments. Due to the large size of the acquired datasets, these images are typically analyzed in HPC clusters. However, local fat nodes for the analysis are also used due to the recent introduction of GPUs. Therefore, the workflow engine must be flexible enough to allow local execution and distributed execution in HPC clusters, which further impose limitations on the use of queues, connection/authentication constraints for accessing the worker nodes, setting up the execution environments on the fly, and so forth.
-
H. Execution with and without graphical capabilities. Graphical interfaces are handy for designing the workflow analysis interactively, making decisions at the sight of the results of the executed steps, including or excluding images from the study, and so forth. However, HPC environments often do not allow graphical interfaces. Therefore, the workflow engine must enable the analysis in these more challenging environments by providing automatic selection tools.
-
I. Flexible visualization. Additionally, different software packages may provide separate data analysis tools for the same entity type. The workflow engine must allow multiple viewers for the same object to coexist.
-
J. User authentication and validation. Another critical issue, especially in shared environments, is the need to restrict access to the data and analysis exclusively to those users allowed to access them. For instance, two different users may share the same workflow engine to analyze their data. Each one must have access to their analysis but should be prevented from accessing the other user’s data.
-
K. Scripting capabilities. On many occasions, it is convenient to define the workflow on-the-fly following some logic that may automatically decide the next step to execute. Scripting languages, such as Python, are very suitable for this kind of application as they combine ease of development with expression richness and an extensive collection of existing libraries. It is desirable that the workflow engine itself could be called from such scripts and grant full access to its functionality through an API (Application Program Interface).
-
L. High performance. The workflow engine should be light in its own computational and storage requirements; that is, the maintenance of the metadata and the status of the execution state of each one of the protocols should not hinder high throughput (e.g., in cryo-EM, the microscope produces a movie of about 8 GB every 5 s and the workflow engine cannot spend a significant part of the computational resources for its function). A similar situation can be caused by the handling of indexes and metadata of millions of images, a common case in cryo-EM, in which thousands of movies are aligned into micrographs containing hundreds of particles each resulting in a few million images.
-
M. Traceability. In cryo-EM, as in many other image-processing domains, the analysis of millions of images must be tailored to the specific structure and acquisition being processed. This involves hundreds of smaller steps, trial and error workflow branches, dozens of decisions, and so forth, until the final result is produced. Keeping track of all the parameters of the hundreds of steps and all the decisions is a formidable task without the help of some specific software. The workflow engine should keep track of all the steps, their inputs and outputs connectivity, and all the parameters used along the path.
-
N. Reproducibility. All steps or even the whole workflow should be reproducible when the same image-processing pipeline is executed twice on the same input data. Some underlying processes hinder this reproducibility with a random initialization, which makes perfect sense from the point of view of image processing. This prevents obtaining the same result if the same program is executed multiple times on the same input. Therefore, the workflow engine should provide the mechanisms to get the same result if the underlying process is deterministic and to compare two different outcomes if it is not.
-
O. Self-reporting. Once an image analysis workflow has been executed, the workflow engine should provide some form of self-report so that the user and an external reader may follow the operations and decision branches to go from the raw input data to the final result. This report should be complete enough to allow a critical assessment of the results without omitting any intermediate step.
-
P. Reusability. Although each project may involve different image-processing steps and user automatic decisions, some parts of the applied workflow could be consistently repeated in other projects. Therefore, a mature workflow engine should allow the definition of templates that can be reused among projects with minimal user intervention. At the same time, it should allow the modification of critical parameters so they can be adapted to the specific project. Ideally, these templates should be publicly available in some repository for more accessible user interchange.
-
Q. Extensibility. The workflow engine must allow extending its operations through some mechanism so that new software packages and functionality can be added without modifying the workflow engine source code.
-
R. Software updates. The workflow engine should provide a mechanism to update itself and the various plugins that perform the image processing as the underlying packages evolve. Some of them make new releases periodically, some others release with irregular time patterns, and some others are never officially released and have to be updated from their source code repository. The workflow engine must identify the need to update, propose to the user to do so, and effectively update and install the different packages. This process has to be done in a vast number of environments (vast concerning visible libraries, compiler and interpreter versions, user permissions, etc.) and must be able to detect and report installation failures.
-
S. Non-restrictive software licensing. The installation of the underlying packages should be as transparent to the user as possible. The free academic use of most software packages allows the workflow engine to automatically download and install most of the different programs. However, some software tools require user registration or are commercial, requiring the user to take specific actions on the web, the command line, or even to install the program by themselves and letting the workflow engine know how to invoke the particular program.
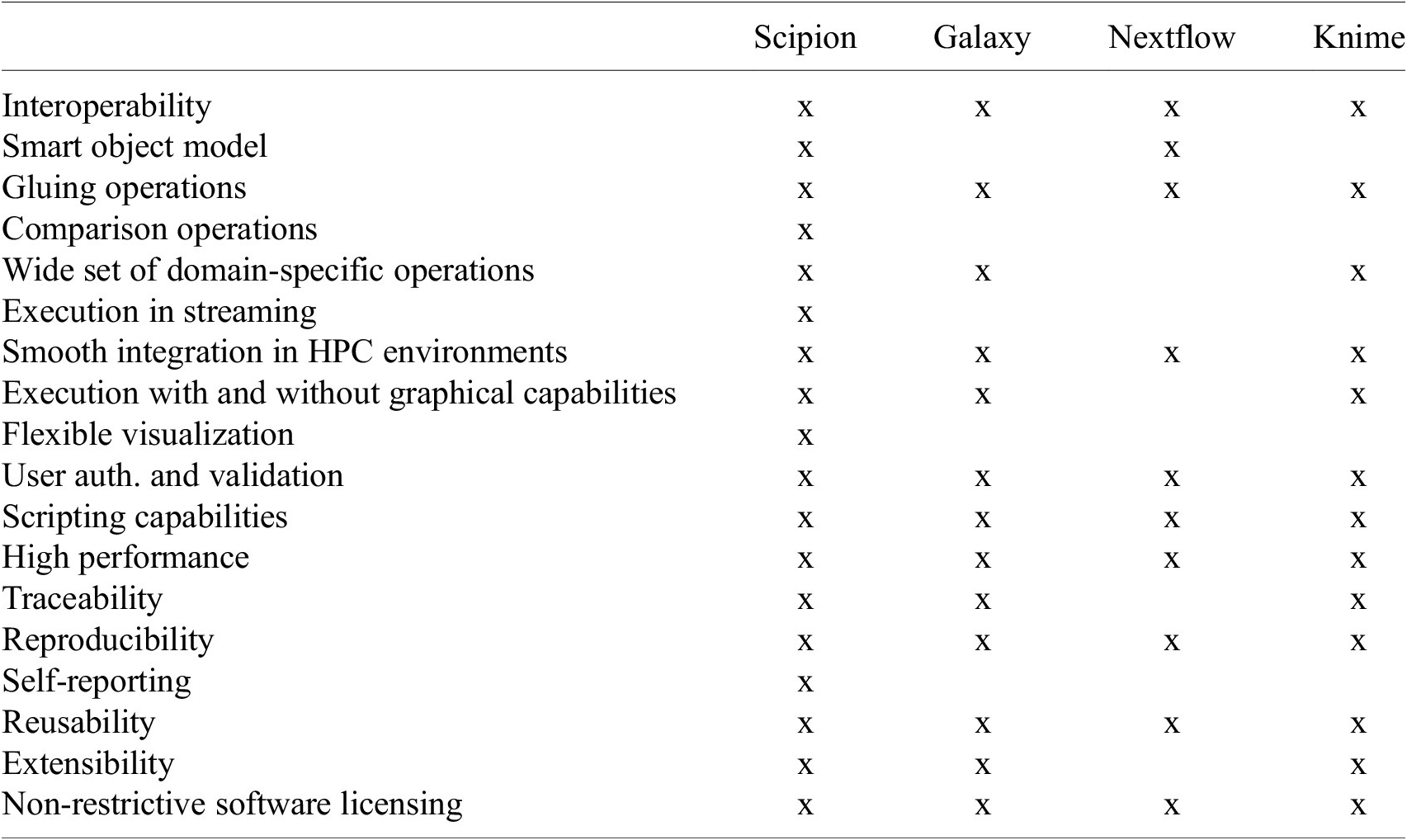
As seen from this long list, designing a workflow engine with all these characteristics is a challenging endeavor. Currently, existing workflow engine solutions typically used in academia or in a bioinformatics setup meet some but not all, simultaneously. Moreover, we understand that some requirements are specific to image processing, some to image processing in streaming with large volumes of data, and others to a complex and evolving software ecosystem. This is the case of cryo-EM, which is why we have developed a workflow engine devoted explicitly to this domain, Scipion. Table 1 compares the fulfillment of all these requirements by some of the workflow engines used in related domains.
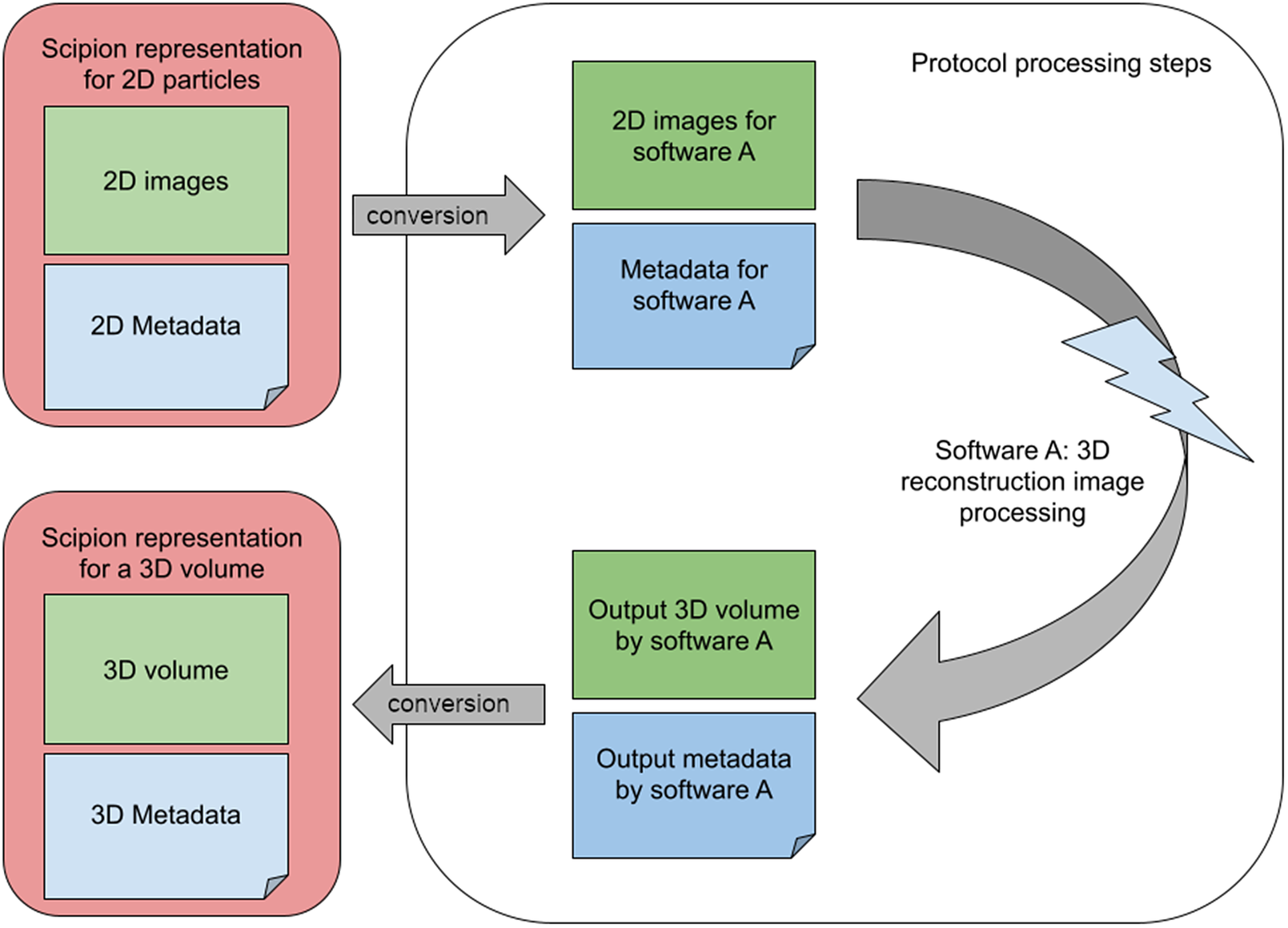
Figure 1. A set of 2D particles in the standard representation must be converted into the internal representation of software X, whose output is collected again by the workflow engine and converted again to its standard representation to serve as the input of further following processes.
Table 1. Table showing how the main functionality Scipion covers is fulfilled by, to the best of our knowledge, some of the workflow engines available to perform similar tasks.
Scipion was first developed in 2013 and has evolved over the years to adapt better and fulfill all above mentioned requirements. Although Scipion started exclusively as a workflow engine for single-particle analysis by cryo-EM(Reference de la Rosa-Trevín, Quintana and Del Cano7), it has been extended, over the years, into other related domains: atomic modeling(Reference Martínez, Jiménez-Moreno and Maluenda8), electron tomography(Reference Jiménez de la Morena, Conesa and Fonseca9), microED(Reference Bengtsson, Pacoste and de La Rosa-Trevin10) and currently, virtual drug screening. It integrates currently over 60 plugins and more than 1,000 protocols, each implementing a high-level task. Scipion has been used in thousands of cryo-EM projects distributed worldwide. At present, we teach four courses a year on image processing using Scipion: (1) single-particle analysis, (2) atomic modeling and flexibility analysis, (3) image processing at the acquisition site, and (4) electron tomography, with a total of hundred attendees per year. Overall, Scipion can be regarded not only as an excellent platform for performing Structural Biology, but also as a suitable engine for any other image or data processing domain with similar requirements to cryo-EM.
In the remainder of the article, we briefly explain the SPA workflow to better illustrate Scipion’s flexibility. Then, we go over all the requirements from a workflow engine and explain how they are met in Scipion. Finally, we provide some insights regarding Scipion’s technical implementation that may be useful in other developments.
2. Single-Particle Analysis by Cryo-EM
A detailed overview of the steps of this image-processing pipeline is explained by Sorzano et al.(Reference Sorzano, Jiménez-Moreno and Maluenda11). In the following, we describe it in detail so that the role of the workflow engine can be understood. First, the electron microscope acquires the so-called movies from a sample. Thousands of these movies are recorded during a session. These movies are made of multiple frames (typically 10–70 frames) whose dose is in the order of 0.5 electrons/Å2 per frame. The signal-to-noise ratio (SNR) per frame is typically around 1/1,000. These multi-frame images have to be aligned into single images (micrographs)(Reference Střelák, Filipovič, Jiménez-Moreno, Carazo and Sánchez Sorzano12). This process accounts for rigid and flexible alignment; after averaging, the SNR rises to 1/100. Then, the microscope aberrations must be estimated from the micrographs; most notably, the defocus, that is, the distance between the sample and the image formation plane, varies from one micrograph to another(Reference Sorzano, Jonic, Núñez-Ramírez, Boisset and Carazo13–Reference Zhang15). To achieve high resolution, this defocus must be determined with a precision below 200–300 Å. Astigmatic micrographs, micrographs for which the defocus cannot be determined, micrographs on the carbon edge, with ice crystals or any other contamination are typically discarded. Even so, many thousand micrographs pass this screening. Then, particles must be identified from these micrographs. Due to the low SNR and contrast, many areas of the micrograph are wrongly identified as particles (see Figure 2). Particles are then classified into allegedly homogeneous groups. This is challenging, as many algorithms tend to favor large, nonhomogeneous groups due to an attraction effect related to the SNR(Reference Sorzano, Jiménez-Moreno and Maluenda5). An initial model is constructed either from the selected 2D classes or the particles being grouped under the selected 2D classes(Reference Vargas, Álvarez-Cabrera, Marabini, Carazo and Sorzano16). Several rounds of 3D classification are now run to identify possible distinct conformations of the macromolecule under study. Finally, once a homogeneous set of particles is achieved, the projection direction of each of the particles is sought, resulting in a final 3D map.
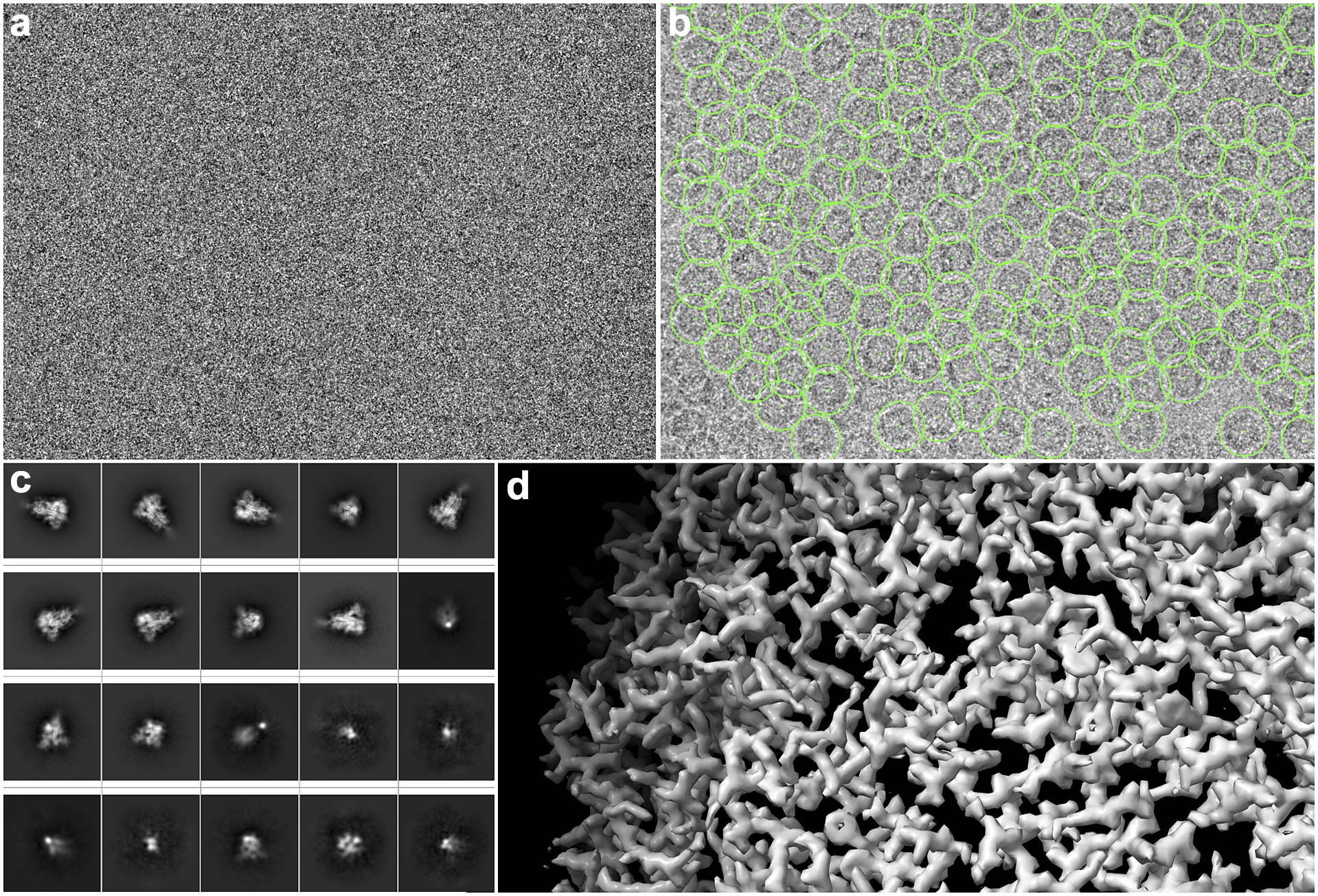
Figure 2. Some images produced during SPA image processing. (a) An example of a section of a movie frame from EMPIAR-10579. (b) Micrograph with hundreds of apoferritin particles with 2D coordinates (green boxes) marked from same EMPIAR dataset. (c) 2D averages of the spikes of SARS-CoV-19. (d) Refined 3D map where the structure of an apoferritin protein can be appreciated.
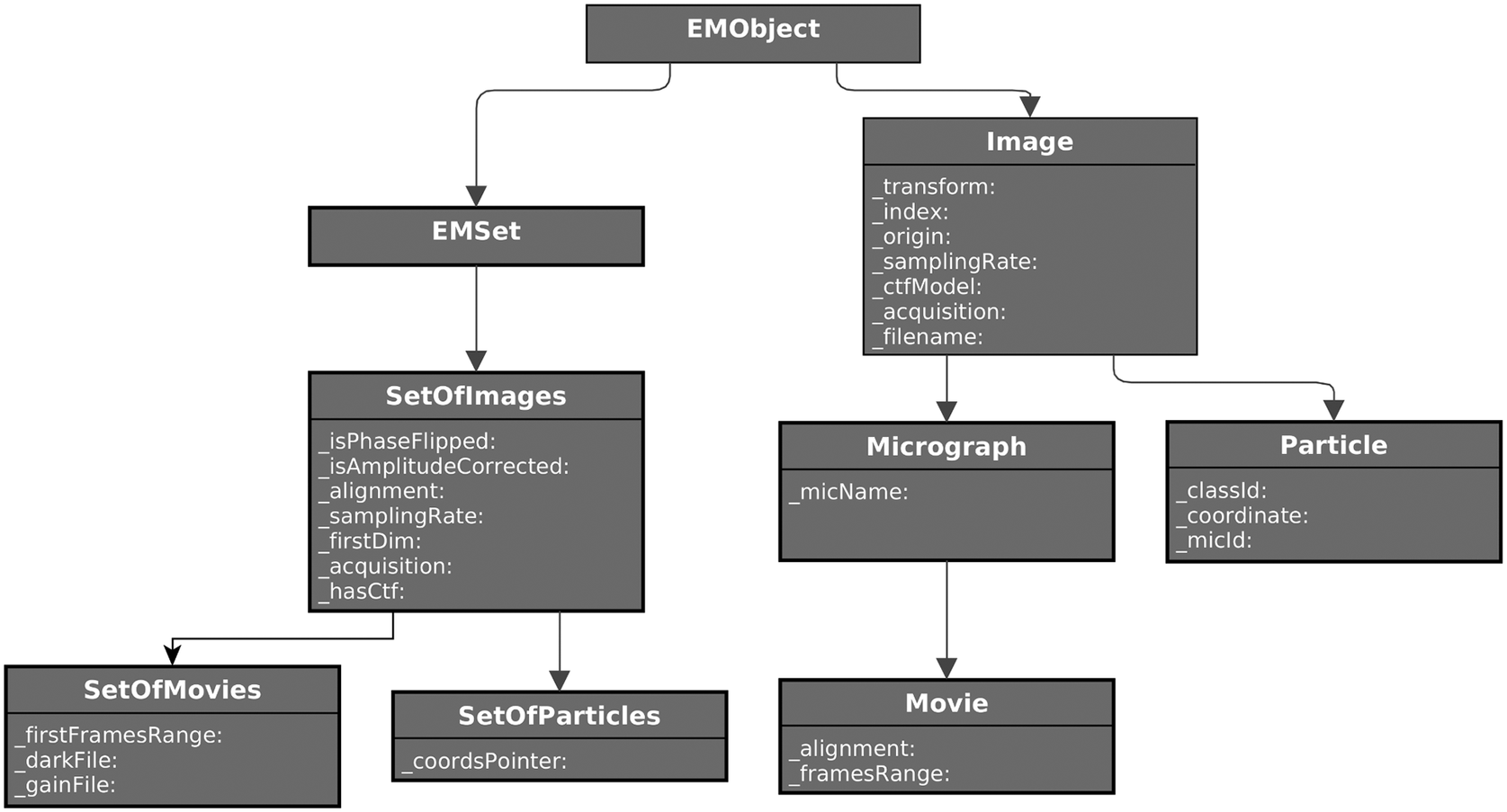
Figure 3. Partial UML diagram of some of the classes defined for SPA image processing. On the left, the set’s hierarchy for images, particles, micrographs, and movies. On the right, single-item classes for the same concepts. Only attributes are shown for clarity.
Due to the low SNR and contrast, all these steps are highly prone to errors. For this reason, each step must be executed more than once, ideally with different algorithms, to identify those parameters that are consistently found. The user defines the degree of consistency, which may vary depending on the dataset and the step within the image-processing pipeline. The simplified description provided in the previous paragraph results in a few hundred steps for a typical project. In this context, the role of the workflow engine is bringing order into the processing, keeping track of everything, allowing interoperability among different algorithms, and guaranteeing the reproducibility of the whole process, among others. In the following section, we describe all the desired requirements for a workflow engine in this domain and how Scipion addresses all the needed issues.
3. Workflow Engine Requirements
In the following sections, we discuss how Scipion addresses the different requirements.
3.1. Interoperability
As in any fast-advancing research field, in cryo-EM, the solutions and software are produced at high speed without waiting for standards to be agreed on. Thus, many image-processing software for cryo-EM have been developed without a coordination effort to unify data and metadata conventions. Most popular software such as Relion(Reference Kimanius, Dong, Sharov, Nakane and Scheres17), Cryosparc(Reference Punjani, Rubinstein, Fleet and Brubaker18), EMAN2(Reference Tang, Peng and Baldwin19), Xmipp(Reference Strelak, Jiménez-Moreno and Vilas20), cistem(Reference Grant, Rohou and Grigorieff21), motioncor2(Reference Zheng, Palovcak, Armache, Verba, Cheng and Agard22), and so forth are able to read and write a common image file format such as mrc, but when it comes to the metadata, they all define their own specification. This is the case for Relion or Xmipp using .star(Reference Hall23) files with their own labels, Cryosparc using NumPy(Reference Harris, Millman and Van Der Walt24) saved files (.cs), or EMAN2 handling a mixture of .hdf, .json, and .box files. In some cases, some of the mentioned software provides metadata conversion methods. However, interoperability among all of them is far from being fully covered, especially when it comes to geometric conventions (the location of the image origin, the direction of the axes and positive rotations, etc.)(Reference Sorzano, Marabini, Vargas, Herman and Frank25). Some of them provide a complete set of methods to cover the whole SPA image-processing pipeline or when missing a step, they integrate third-party software to cover it. This is the case for Relion integrating cistem’s CTF estimation or Cryosparc integrating Topaz(Reference Bepler, Morin and Rapp26). If the data are “friendly,” image processing may go perfectly fine doing it all inside one of those “complete” software. If the results are not satisfactory, users may want to try another software. In this case, things can get complicated. If the software, the user wants to try, does not provide an entry point for importing current processed data and metadata, the user needs to convert them. While converting data (image files) is probably the most straightforward task, metadata conversion is the most challenging one. Most likely, the metadata’s elements described range from thousands to millions. Therefore, users may not go for a manual approach. Scripting becomes crucial at this point. Additionally, metadata may store complex information in different conventions. For example, description in SPA of the alignment information (shifts and rotation). Relion stores this information in two or three shift fields plus three fields for the rotation (three Euler angles in Z, Y, Z). EMAN2 uses Euler angles but uses the Z, X, Z’ convention. Proper conversions require understanding an Euler matrix and the exact conventions followed by each software the user may want to use. Even if the user has the skills and time to implement them, there is a high risk of making errors in the process. And, in the ideal case of no mistakes being committed, annotating all the steps for a future repetition or simply reporting the steps in a future scientific article in an attempt to follow the FAIR principles(Reference Wilkinson, Dumontier and Aalbersberg27) becomes an issue.
3.2. A smart object model
Scipion integration is firmly based on converting one software’s output into another’s input. For this, data and metadata have to be correctly converted. Scipion does not provide multiple conversions, for example, from software A to software B and C, from software B to A and C, and from software C to A and B, which means six conversions for three software, or more generically, N*(N – 1), being N the number of software integrated. On the contrary, Scipion defines a standard object model. This is a set of Python classes that can hold any data and metadata produced by any integrated software at a specific step. Therefore, software A’s output is converted to Scipion’s standard object model. If another software needs to consume this output later, it reads the Scipion object model instead of the software’s A output. Conversions follow the following pattern: Software A produces its output A’, which is converted to Scipion standard object model S′, which is finally converted to software B input B′. More generically, the amount of conversions needed is N*2. In addition, Scipion provides generic self-persistence data types like string, integer, float, lists, object, and so forth. These “primitive” Scipion data types allow the definition of more complex objects to model any output. This has been the case for SPA image processing, where we have defined specific objects such as micrographs, movies, CTFs, FSCs, particles, classes2D, coordinates2D, volumes, and so forth. All these objects in combination with a set object aiming to store collections of previous elements provide a stable model capable of storing all data and metadata produced during the SPA image processing. In the few cases where the standard model is insufficient, objects are automatically extended with specific attributes that are also automatically persisted. Those extra attributes are ignored by other software but probably read later by the same software that extended the object.
The objects that flow through the image-processing pipeline have to be persisted on disk so that they can be recovered later. This is achieved in Scipion by translating this flexible object data model into a set of SQLite tables. In this way, each set has its SQLite file, and we avoid having a single table with millions of entries corresponding to the many millions of images handled along the processing. This implementation allows us to speed up the reading/writing of objects and also to isolate possible database corruption problems.
Defining the standard object model for a specific domain (in this case, single-particle analysis) is not trivial. Therefore, it may take time to get a mature model, but at the same time, if versatile enough, it could be a starting point for a future standard definition initiative the field may dive into (see Figure 3).
3.3. Gluing operations
When carrying out a complex analysis of millions of images to generate one or several maps according to the different molecular conformations present in the sample, we can distinguish two different types of operations: (1) “heavy” operations performing the domain-specific tasks on thousands or millions of images (align thousands of movies into micrographs, estimate their microscope aberrations, identify particles, align those particles, etc.), and (2) “light” operations performing house-keeping tasks (divide large sets into smaller sets for comparison purposes, select a subset of images according to some criteria, filter out images that do not meet the required quality, merge subsets into larger sets, combine the information of multiple algorithms computed on the same set, compute a subset of a more extensive set according to the information provided by another subset, etc.). All these housekeeping operations are crucial for successfully completing a project and the workflow engine must support them. One of the most challenging tasks is to keep track of the information of a particle, even if it is spread over multiple objects. In Scipion, we assign each particle a ParticleID consistent across the project. An interesting feature of our implementation is that the ParticleID represents the information of a particular location within a micrograph, irrespective of the downsampling and possible transformations that the image could suffer. Also, our domain has a natural hierarchy of images: a single micrograph usually contains many different particles. This hierarchy has to be kept along the process so that the MicrographID corresponding to a given ParticleID is always known. This information allows performing subsets of micrographs according to some criteria and then subsets of particles coming from a given subset of micrographs.
3.4. Comparison operations
Image processing in SPA involves the estimation of many different parameters: whether a location in a micrograph represents the central point of a macromolecule (a binary parameter indicating whether a coordinate points to a particle or not), the three-dimensional orientation of a given particle (a categorical parameter with several possible values), the defocus and alignment of that particle (some continuous parameters), and so forth. The different algorithms performing the “heavy” workflow operations aim to estimate these parameters. However, they have to do it in an environment where the SNR is around 1/100, meaning there is about 100 times more noise than signal. Consequently, these estimations are prone to mistakes(Reference Sorzano, Jiménez-Moreno and Maluenda5), and it is crucial to provide quality control operations. These quality control algorithms may be addressed at (a) the images themselves, assigning a quality score that the user may use to filter out the images (algorithms such as xmipp screen particles(Reference Vargas, Abrishami and Marabini28), relion 2D class ranker(Reference Kimanius, Dong, Sharov, Nakane and Scheres17), and xmipp movie gain(Reference Sorzano, Fernández-Giménez and Peredo-Robinson29)); (b) checking that the estimated parameters are within a given range (for instance, the defocus, the correlation with some reference images, etc.); and (c) comparing the estimates of two equivalent algorithms and checking that their difference is smaller than a given threshold (estimates like angular assignment, defocus, etc.). Additionally, users may want to try different attempts and modify some quality thresholds to check if the results can be improved. All these trials must be traced and should be reproducible, and again, the help of the workflow engine in this regard is invaluable.
3.5. Wide set of domain-specific operations
Besides the good technical features of a workflow engine, a crucial aspect of its success in a given domain is the availability of operations for a specific domain. Galaxy(Reference Jalili, Afgan and Gu30) is an example of a well-known workflow engine for genomics and related-omics analysis. It performs over 1,000 operations in these domains. However, it does not offer any tool in cryo-EM image processing, and very few for atomic modeling. In this regard, Scipion offers over 1,000 operations in Structural Biology around the following domains: single-particle analysis, electron tomography, atomic modeling of cryo-EM maps, and virtual drug screening. The availability of so many operations in a specific domain dramatically facilitates the analysis of complex data, even in cases where a relatively rare operation is needed. Furthermore, it becomes a platform for developers to integrate the developed tools. This is especially useful for those developers whose software packages only cover part of the image-processing pipeline.
3.6. Execution in streaming
Cryo-EM image acquisition produces images (uncorrected motion movies), and its quality assessment is only possible by accomplishing a few image-processing steps in the pipeline. An electron microscope produces a movie every 10–60 s and acquisition time typically lasts 1 or 2 days at a cost of 500–5,000 euros/day depending on the subsidies provided by the host institution(31, Reference Cianfrocco and Leschziner32). Thus, microscope users need to assess the quality of the new data as soon as possible to make corrections on the ongoing acquisition process if data does not reach quality standards. Scipion protocols allow real-time data assessment working in “streaming” mode. A protocol can be launched without having all input data available. It will actively look for new input during its execution or stay idle until it checks again. If further input elements are discovered, they are processed, and new results are added to the protocol output. This process lasts until all the input data is processed and the input is closed (no more information will appear).
3.7. Smooth integration in HPC
HPC clusters are common computing resources in academic institutions. They usually provide access to the cluster through one or more “login nodes.” Processing commands are launched from the login node to a job scheduling system(33) like Slurm(34), Torque(35), or others. Scipion can be configured to run with job scheduling systems through a dedicated configuration file(36). In this way, Scipion can use the existing computational infrastructure and resources. In addition, modern cloud infrastructure like Amazon web services, Microsoft Azure, or other academic clouds can also be used to deploy Scipion to them(37). Scipion has also been successfully containerized in docker instances(38) with full GPU integration.
3.8. Execution with and without graphical capabilities
Graphical interfaces are handy to monitor the correct execution of the image-processing workflow, identify particles in a micrograph, make decisions on which images to include in the analysis, filtering out images based on the empirical distribution of a given quality measure, and make subsets based on image quality or similarity. However, there are situations where graphical interfaces are not allowed, as in some HPC environments, or are not convenient. In these situations, the workflow engine must be able to execute the workflows without any graphical interface or even not loading any graphical library, which may not be available. Then, operations typically performed by a user (identifying particles, quality control of the process, etc.) must be performed automatically by some of the protocols. The introduction of deep learning algorithms for particle picking, such as Cryolo(Reference Wagner, Merino and Stabrin39) or Topaz(Reference Bepler, Morin and Rapp26), has enormously simplified this task, which used to require a significant amount of user intervention. Automatic quality control is more complicated, although some advances have been put forward in this regard(Reference Sorzano, Jiménez-Moreno and Maluenda11), and it is an active research topic.
3.9. Flexible visualization
Each of the steps of the pipeline produces one or more outputs. Scipion provides a mechanism to register viewers for specific types of outputs. A viewer is a piece of code capable of visualizing data of a given type within the smart object model described in requirement B. It usually wraps existing viewers provided by the integrated software. For instance, reconstructed maps can be sent to the ChimeraX(Reference Pettersen, Goddard and Huang40) or the default map visualization utility of Xmipp, Bsoft(Reference Heymann and Belnap41), or EMAN2(Reference Tang, Peng and Baldwin19). Most of the outputs produced by the protocols have one or more viewers able to display them. Additionally, a specific protocol could define a list of results supporting the execution with plots or images. In more complex cases, viewers can prepare the results, apply some statistics or create complex animations to provide users with a complete set of analysis tools (see Figure 4).
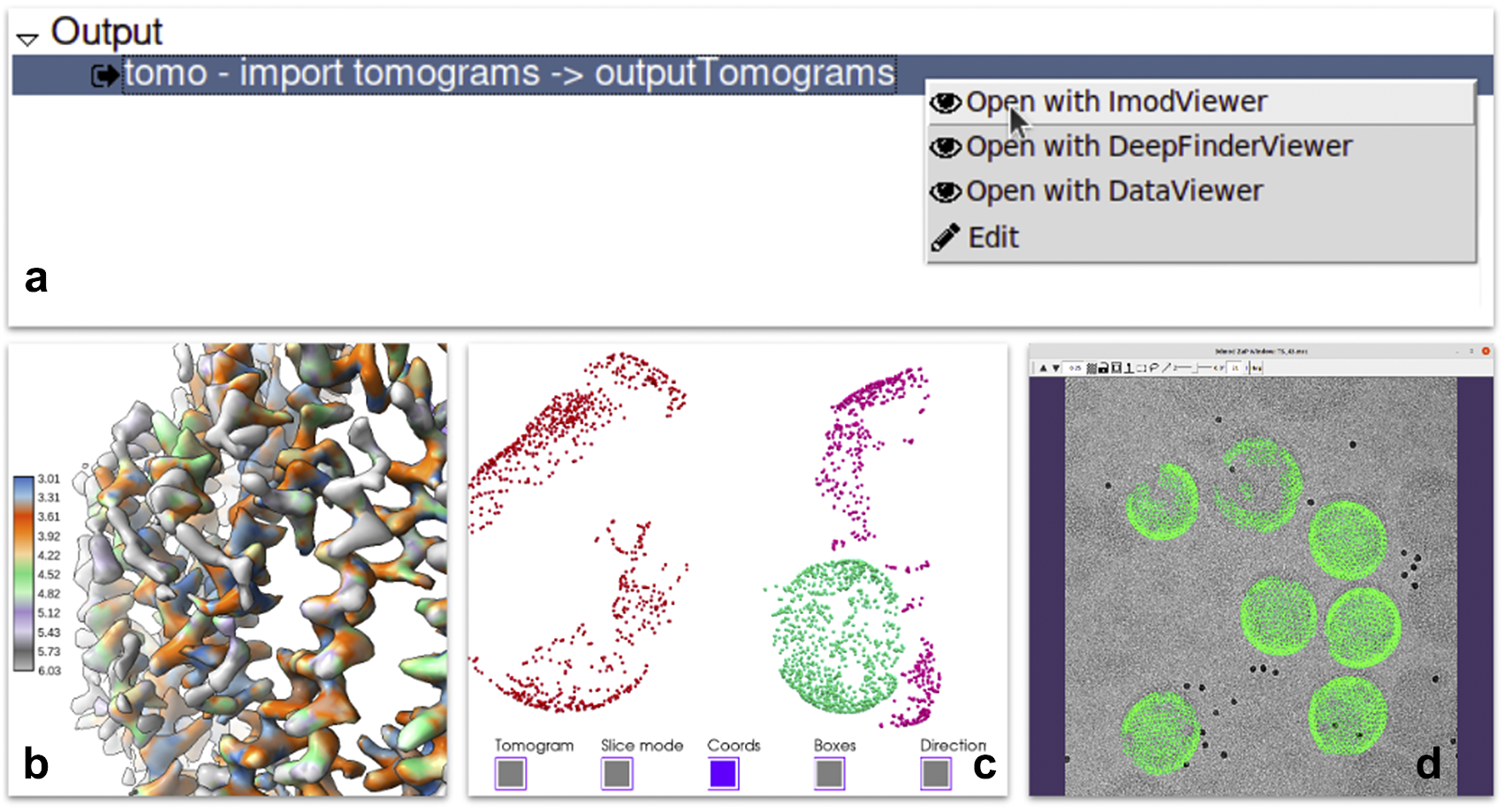
Figure 4. Some of the visualization tools available in Scipion. (a) Context menu offered when right-clicking in a set of tomograms displaying three different viewers: Imod, Deepfinder and Dataviewer (Xmipp). (b) Local resolution results produced by Xmipp monores(Reference Vilas, Gómez-Blanco and Conesa42) and shown in ChimeraX(Reference Pettersen, Goddard and Huang40). (c) 3D coordinates picked on a tomogram generated by pySeg(Reference Martinez-Sanchez43) and shown in Tomoviz plugin. (d) 3D coordinates projected on the corresponding tilt series shown as fiducials in Imod’s fiducial viewer as a way to visually verify all metadata and data looks correct after a “relion4 prepare” process to enter subtomogram averaging.
3.10. User authentication and validation
Scipion can be installed in a private machine that can be accessed only by a single person or in a shared computer or cluster accessed by several users. However, access to the data should be restricted to the user that created it or the users with whom the owner decides to share it. Scipion is an application installed locally instead of a web application with remote access. Consequently, we delegate user authentication and data privacy to the standard Unix mechanisms (Unix user authentication and file/directory access permissions). In this way, the data can be defined by the user to be only visible to herself, by a set of Unix user groups, or by any user of the computer. Some groups create an image-processing user shared by all the lab members, but this practice is entirely discretionary to the policy of each team.
3.11. Scripting capabilities
An essential feature of modern workflow engines is allowing programmatic access to the image-processing workflow. Scipion is implemented in Python and exposes all its internal functionality through an API that can be accessed at https://scipion-em.github.io/docs/release-3.0.0/docs/api/scipion-API.html. Through the API, an external script can control the creation of a project, the creation of the different protocols one-by-one and their interconnection (which outputs are the inputs of any of the protocols), all protocol parameters, and so forth. It might be useful in scenarios where we want to automate several tasks without user intervention. We may also create the workflow taking decisions within the script. For example, depending on the number of particles available, we may decide to split the set of particles into smaller subsets for faster processing.
3.12. High performance
Scipion does not perform any of the image-processing tasks by itself. Instead, it relies on the underlying software packages to perform these operations. However, the inputs of a protocol must be converted into the data and metadata formats required by the underlying program. The program outputs must be transformed back to Scipion’s representation of the object types produced. These conversions must add minimal overhead on the actual image-processing time so that the workflow engine does not add any significant delay in the image-processing pipeline. In Scipion, we have paid attention to these issues trying to make the conversions as efficient as possible.
High performance applies also to any Scipion’s native visualization. This is particularly important for visualizing sets of particles, which may contain millions of images. In this case, an efficient display strategy is adopted in which we only need to read those images displayed at any given moment, usually a few tens.
3.13. Traceability
Due to the natural complexity of the workflow, especially in challenging cases, users may want to try several analysis alternatives, combine software, and add validation methods in between. Annotating all the decisions taken with the purpose of later reporting or reproducibility becomes a critical and highly tedious task. Traceability is one of Scipion’s core principles, and it comes out of the box when using it. Each protocol in the pipeline is represented in Scipion as a single rectangle in the main project window. The protocol stores all the parameters used and allows their edition in a dedicated form (Figure 5). The whole workflow can be exported as a JSON file containing all the information and sent to EMPIAR(Reference Iudin, Korir, Salavert-Torres, Kleywegt and Patwardhan44), a repository of raw data for cryo-EM. EMPIAR understands this Scipion JSON file and represents it on the web using a web component developed by us(45), as shown in some entries (EMPIAR-10891, EMPIAR-10516, and EMPIAR-10514).
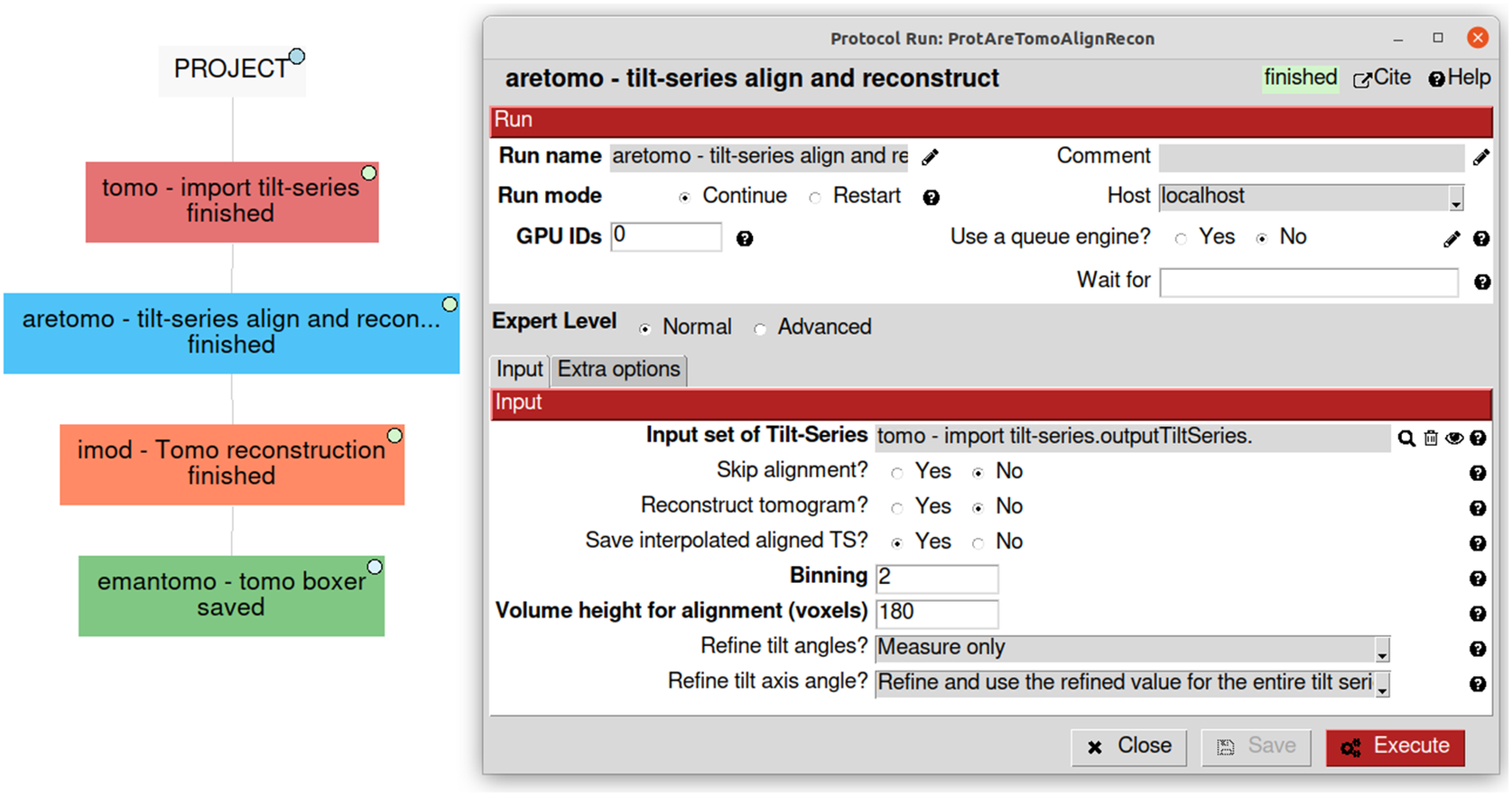
Figure 5. Simplified visualization of some possible first steps of a tomography workflow showing a use case of four different software on the left. On the right, an example of the “aretomo—til-series align and reconstruct” protocol detailing its parameters and its link to the “tomo—import tilt-series” output called “outputTiltSeries.”
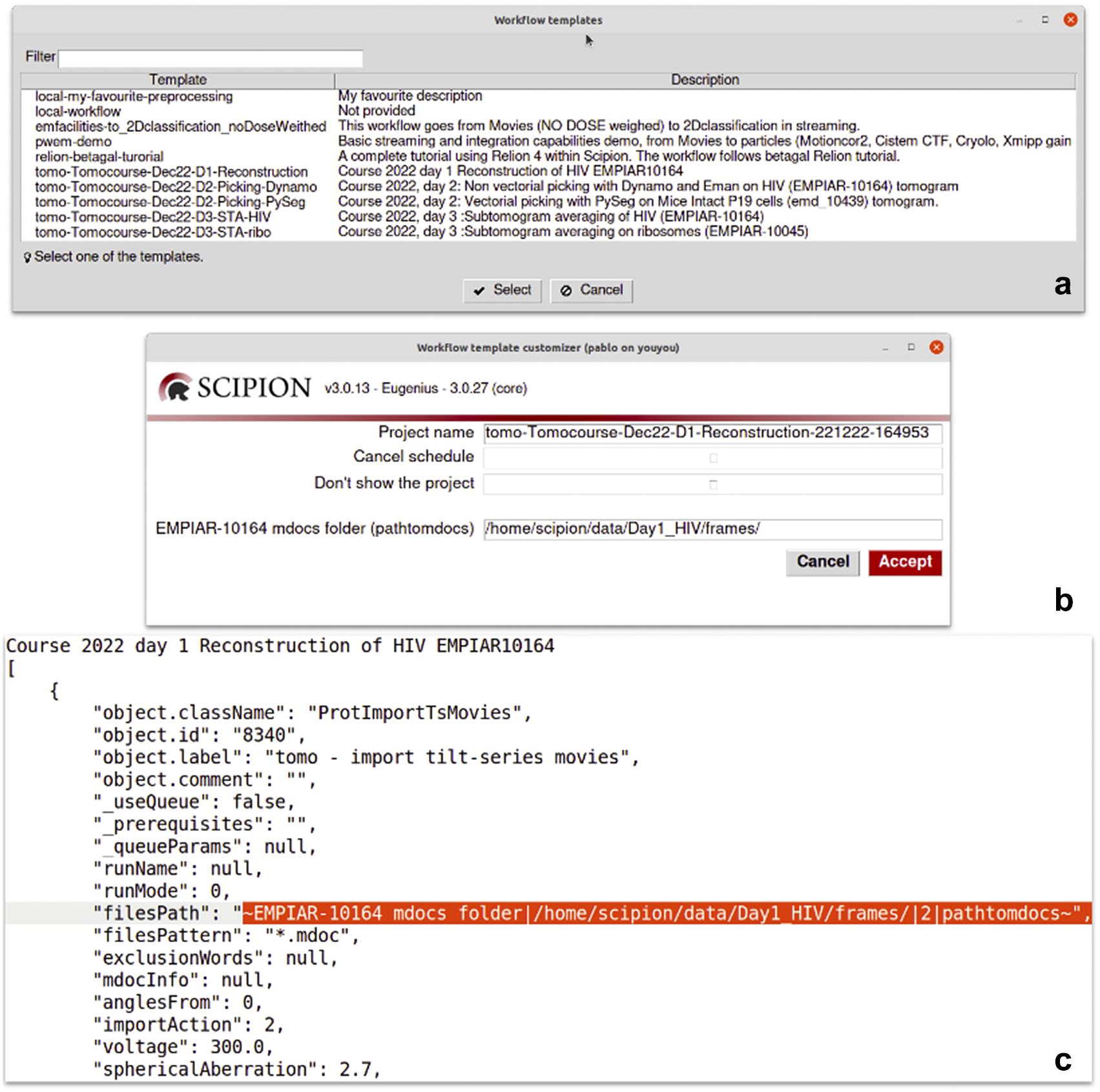
Figure 6. Details of the dynamic templates. (a) List of templates found in a Scipion installation. Those starting with “local-” are user templates found in a dedicated folder for templates. The rest are provided by some of the plugins like relion or tomo plugin. (b) A dynamic window is shown after selecting the “Tomocourse-Dec22-D1-Reconstruction” template. It offers to choose the name of the project to be created and to cancel scheduling or avoid showing the project once created. For this particular case, it additionally shows one dynamic field to be asked for its value: the “EMPIAR-10164 mdocs folder” field. (c) Excerpt of the same template opened in a text editor showing the part where the dynamic field is defined (filesPath).
3.14. Reproducibility
The accurate and detailed recording of algorithm parameters, inputs, and outputs of all the image-processing steps performed along the pipeline enables launching each one of the steps with exactly the same inputs and parameters used in the execution process, leading to the final result deposited in the public databases. In this way, the whole workflow can be exactly reproduced from the beginning to the end as long as the underlying algorithms are deterministic. Nevertheless, reproducibility is limited by the underlying software packages and hardware. Particularly, some algorithms start from a random solution that is later refined. If this random initial solution makes the algorithm not deterministic, Scipion cannot guarantee to obtain the same results. Also, we have observed that deterministic algorithms running on two different computers may yield different results. This result comes from a combined influence of the underlying hardware and the system libraries installed.
3.15. Self-reporting
Due to the complexity of the image analysis performed in cryo-EM, the final result is never the “direct” processing of all the movies acquired. Instead, to reach these results, dozens of decisions have been taken along the path (particles selected and discarded in the analysis, conformational class of each particle and its relative orientation with respect to it, parameters of each of the image-processing algorithms employed, etc.) Without this information, it is impossible to fully understand all the steps leading from the raw acquired data to the final result. This information is never reported in scientific papers to a level of detail that allows reproducibility or even its complete understanding. In Scipion, we have developed a tool that uploads a detailed summary of the executed workflow to a server(46). Although recommendable, this uploading is optional.
3.16. Reusability
Scipion workflows can be exported to serve as templates for future analyses. This is particularly relevant in the first steps of the processing, where few decisions are taken, or in specific parts of the analysis where the sequence of steps is always the same. We provide a repository of valuable workflow templates at http://workflows.scipion.i2pc.es/ as JSON files. Users can import those templates into existing projects or create new projects based on a template. An imported workflow template will likely have some parameters specific to each project that need to be set up before running any protocol. This is the case for most of the session’s acquisition parameters (path to images, sampling rate, electron dose, voltage, particle size, and symmetry). Those parameters may be spread throughout the workflow. Since reviewing all the protocols to update those parameters could be tedious, with the risk of running the complete workflow with the wrong parameters, Scipion provides dynamic templates(47). Dynamic templates are also JSON files, similar to the already mentioned workflow files, but with some parameters marked. When importing a template, Scipion looks for these special marks. If found, it will pop-up a window with all the marked parameters in the dynamic template before creating and running the workflow. This functionality allows users to fulfill parameters changing from one dataset to another, adapting the workflow template to the specific data being processed (see Figure 6). Alternatively, the parameters can also be given through the command line without needing any graphical interface.
3.16.1. Designing, scheduling and queuing
Scipion also allows the user to design a workflow in advance without running it. Protocols can define in advance their output type and name. This is enough for other protocols to be linked. At this initial stage, their inputs are not ready and cannot be executed but can be scheduled. A scheduled protocol is run in a separate process that keeps checking if everything is ready to run. Once a scheduled protocol finds all its inputs available, it launches itself to start its actual image processing. Optionally, a protocol can be told to wait for the completion of another “unrelated” protocol before starting, even in the absence of any input dependencies. This “wait for” parameter (see Figure 5) is checked during the scheduling process and stops the protocol from being executed until the dependency is satisfied. Scheduling will enable users to avoid idle CPU/GPU time in machines and plan and schedule an overnight workflow to keep processing machines busy at night.
3.17. Extensibility
Software packages and new operations can be added to Scipion through plugins. A Scipion plugin (https://scipion-em.github.io/docs/release-3.0.0/docs/developer/creating-a-plugin.html) is a Python package that follows some specific conventions. Beyond these requirements, the Python module defines its structure. Once installed, Scipion is capable of discovering all the new protocols and new data types defined by the plugin so that they become immediately available to the user. Protocols are wrappers to the underlying image-processing algorithms. Typically, they extract the information from the inputs to the protocol, convert it to the format required by the underlying software, execute it, and send the output back to Scipion’s representation. This plugin structure makes Scipion very flexible to cover all the processing in single-particle analysis, electron tomography, atomic modeling, virtual drug screening, and chemoinformatics(48).
3.18. Software updates
Scipion plugins are generally distributed through PyPi (https://pypi.org/search/?q=scipion), although other installation possibilities are also supported. Every time Scipion is launched, it checks for updates and informs the user whether there is an update of the workflow manager. Plugins update status can be checked in the plugin manager. Once a plugin is updated, its binaries (third-party software it integrates) may also be updated in case new releases are available. This way, we can keep the most up-to-date versions of the underlying programs.
3.19. Software licenses
Most scientific packages integrated into Scipion do not have any special software license or are free for academic use. A few require user registration or a specific installation procedure involving some form of license. In those cases, where the installation can be performed without special permission, Scipion handles all the processes of downloading the software, compiling it if necessary, and installing it in an accessible path. In those cases, where the license is granted by filling out a form, Scipion guides the user to that form and then handles the installation. For commercial software or packages that require a particular installation procedure, users can install the program by themselves and simply inform Scipion about its location. This possibility is available to all software packages and is very useful when working with their development versions.
4. Conclusions
Within the image-processing community, researchers usually center on the mathematical and algorithmic aspects of the computational solutions to the various image-processing tasks. They normally focus on one specific problem (image alignment, denoizing, restoration, reconstruction, etc.) and devise sophisticated algorithms to tackle it. Although this approach is crucial for solving the “low-level” tasks at the core of the whole image-processing activity, hundreds of these low-level operations are sometimes needed to solve a biological macromolecule’s three-dimensional structure from the raw acquired images. Moreover, due to the extremely low SNR, the underlying algorithms are prone to errors in estimating the various parameters involved. The only way of identifying these mistakes is by estimating the same parameter with different algorithms and comparing their results, but this multiplies the number of low-level operations at least by a factor of 2. If the various estimates are close, averaging them should give an even better estimate. If the estimates are far from each other and some cluster, then probably that is the area of the actual parameter. If they do not cluster, the corresponding image is too noisy to allow a reliable estimation of its pose or conformation.
Tracing the decisions and parameters of these hundreds of operations is critical for the reproducibility and accountability of the final result. Interoperability between the different software packages is also essential to allow the comparison of other estimates of the same parameters. It is, at this point, where workflow engines come into play. They help to solve high-level problems. Whereas most workflow engines usually concentrate in one application domain, Scipion does in image processing by cryo-EM and some other related topics (atomic modeling and virtual drug screening). This application domain has some unique features that are not shared with other domains, for instance, the large size of the data to process, defining data types so that the information has well-defined semantics, the need to process as the data are acquired, and the need to call locally installed programs with very different software dependencies and execution environments. On the other hand, it has needs that are shared with other domains. In particular, most workflow engines must be efficient, integrate into HPC environments, extendible, traceable, and reproducible, have a data privacy policy, and so forth.
Overall, Scipion is an excellent environment for the execution of workflows with similar characteristics to the ones exposed in this article (large volumes of data, strong semantics, streaming, and command-line programs with long execution times). It is also an excellent environment for any other scientific domain related to Structural Biology or Structural Bioinformatics. Complex image analyses beyond cryo-EM also fall into the first category, and some groups are extending Scipion to work with fluorescent microscopy data.
Scipion’s first release (1.0.0) was in February 2016. Since then, thousands of image-processing projects (https://scipion.i2pc.es/report_protocols/projectStats) have been created and thousands of installations (https://scipion.i2pc.es/report_protocols/installationStats) have been performed.
All software code, Scipion core and plugins, is open-sourced and publicly available at https://github.com/scipion-em.
Acknowledgments
The authors would like to thank the more than 60 developers (https://scipion.i2pc.es/acknowledgements) that have contributed to either developing the core or any of the plugins evidencing that Scipion open-source project is supported by numerous developers in the community.
Authorship contribution
Y.C.F., J.J.M., G.S., J.M.R.-T., R.M., C.O.S.S., and P.C. have contributed to Scipion core and cryo-EM domain extension. A.C., B.R.F., L.C., M.G., M.I., M.M., P.L., R.M., and I.S. have beta tested all the functionality covered by Scipion, not only SPA but modeling and tomography They also have assisted in the virtualization of Scipion and in all the training courses we yearly organize. M.M. and R.M. have extended Scipion toward modeling. A.G.M. and D.M. have contributed to making streaming functionality better and more robust not only for SPA but also for tomography. D.H. has extended Scipion to the cheminformatics domain. D.H., E.F.-G., F.P.I.-G., and J.L.V. have extended Scipion toward cryo-EM tomography. D.S. has made exhaustive and critical performance analyses. E.R.-A. has improved the SPA tools in Scipion for local resolution analysis and sharpening tools. J.K. and D.H. are extending Scipion toward SPA flexibility analysis. J.M.C. and C.O.S.S. are leading the project and providing scientific advice.
Competing interest
The authors declare no competing interests exist.
Funding
The authors acknowledge the economical support from MICIN to the Instruct Image Processing Center (I2PC) as part of the Spanish participation in Instruct-ERIC, the European Strategic Infrastructure Project (ESFRI) in the area of Structural Biology, Grant PID2019-104757RB-I00 funded by MCIN/AEI/10.13039/501100011033/ and “ERDF A way of making Europe,” by the “European Union.” European Union (EU) and Horizon 2020 through grants: EOSC Life (INFRAEOSC-04-2018, Proposal: 824087), HighResCells (ERC – 2018 – SyG, Proposal: 810057), and iNEXT-Discovery (Proposal: 871037).









