Article contents
Selective sweep and the size of the hitchhiking set
Published online by Cambridge University Press: 01 July 2016
Abstract
Just after the fixation of an advantageous allele in the population (this spread is called a selective sweep), the neutral genes close to the site under selection tend to have the same ancestor as the gene under selection. However, some recombinations may occur during the selective sweep and break the link, which reduces the number of hitchhiking alleles. We consider a large selection coefficient α and extend the results of Etheridge, Pfaffelhuber and Wakolbinger (2006) and the work of Pfaffelhuber and Studeny (2007) about genetic hitchhiking, where the recombination rate scales with α/log α. We first describe the genealogy at an arbitrary number of partially linked neutral loci, with an order of accuracy of 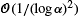 in total variation. Then, we use this framework to obtain an approximate distribution for the size of the hitchhiking set at the end of the selective sweep, with the same accuracy.
in total variation. Then, we use this framework to obtain an approximate distribution for the size of the hitchhiking set at the end of the selective sweep, with the same accuracy.
- Type
- General Applied Probability
- Information
- Copyright
- Copyright © Applied Probability Trust 2009
References
- 5
- Cited by


