Introduction
Myxosporeans are cnidarian parasites with a complex 2-host life cycle that involves annelid worms as definitive hosts and usually fish as intermediate hosts (Lom and Dyková, Reference Lom and Dyková2006). Several myxosporeans cause severe fish diseases that result in considerable economic losses in the fishing and aquaculture industries (Pote et al., Reference Pote, Hanson and Shivaji2000; Kent et al., Reference Kent, Andree, Bartholomew, El-Matbouli, Desser, Devlin, Feist, Hedrick, Hoffmann, Khattra, Hallett, Lester, Longshaw, Palenzuela, Siddall and Xiao2001; MacKenzie et al., Reference MacKenzie, Kalavati, Gaard and Hemmingsen2005; Feist and Longshaw, Reference Feist, Longshaw and Woo2006). The genus Henneguya Thélohan, 1892 of the family Myxobolidae Thélohan, 1892, is one of the most diversified myxosporean taxa, comprising more than 250 species described worldwide (Rangel et al., Reference Rangel, Santos and Rocha2023). Henneguya are predominantly histozoic parasites of freshwater fishes, occurring less frequently in brackish/marine hosts. The gills are the main site of infection for these parasites, but they have also been reported from the epidermis, heart, kidneys, intestine, fins, urinary bladder, brain, eyes, gallbladder, liver, gonads, swim bladder, abdominal cavity, cartilage, spleen, among other organs and tissues (Eiras, Reference Eiras2002; Eiras and Adriano, Reference Eiras and Adriano2012; Rangel et al., Reference Rangel, Santos and Rocha2023).
The European seabass, Dicentrarchus labrax (Linnaeus, 1758) (Moronidae), is an important commercial fish intensively cultured in Portugal and in the Mediterranean (Rocha et al., Reference Rocha, Cabral, Marques and Gonçalves2022). Several myxosporeans have been described or reported from this fish host: Sphaerospora testicularis Sitjà-Bobadilla and Álvarez-Pellitero, Reference Sitjà-Bobadilla and Álvarez-Pellitero1990 from the testis; Kudoa dicentrarchi (Sitjà-Bobadilla and Álvarez-Pellitero, Reference Sitjà-Bobadilla and Álvarez-Pellitero1992) from the connective tissue of all organs; Ceratomyxa labracis Sitjà-Bobadilla and Álvarez-Pellitero, Reference Sitjà-Bobadilla and Álvarez-Pellitero1993 and Ceratomyxa diplodae Lubat, Radujković, Marques and Bouix, Reference Lubat, Radujković, Marques and Bouix1989 from the gallbladder; Ortholinea labracis Rangel, Rocha, Casal, Castro, Severino, Azevedo, Cavaleiro and Santos, Reference Rangel, Rocha, Casal, Castro, Severino, Azevedo, Cavaleiro and Santos2017 from the urinary bladder and a Myxobilatus sp. from the kidney and urinary bladder (Lubat et al., Reference Lubat, Radujković, Marques and Bouix1989; Sitjà-Bobadilla and Álvarez-Pellitero, Reference Sitjà-Bobadilla and Álvarez-Pellitero1990, Reference Sitjà-Bobadilla and Álvarez-Pellitero1992, Reference Sitjà-Bobadilla and Álvarez-Pellitero1993; Santos, Reference Santos1996; Rocha et al., Reference Rocha, Rangel, Castro, Severino, Azevedo, Santos and Casal2016; Rangel et al., Reference Rangel, Rocha, Casal, Castro, Severino, Azevedo, Cavaleiro and Santos2017; Casal et al., Reference Casal, Soares, Rocha, Silva, Santos, Nascimento, Oliveira and Azevedo2019). Except for the latter, all these species have been reported to occur in stocks of D. labrax cultured in southern Portuguese fish farms (Rangel et al., Reference Rangel, Castro, Rocha, Severino, Casal, Azevedo, Cavaleiro and Santos2016, Reference Rangel, Rocha, Casal, Castro, Severino, Azevedo, Cavaleiro and Santos2017; Rocha et al., Reference Rocha, Rangel, Castro, Severino, Azevedo, Santos and Casal2016; Castro et al., Reference Castro, Cavaleiro, Rangel, Rocha, Severino, Casal and Santos2018). Two other myxosporeans, originally described from fish other than European seabass, have also been reported from this host in other geographic locations: Enteromyxum leei (Diamant, Lom and Dyková, 1994) in the gut (Palenzuela et al., Reference Palenzuela, Redondo and Álvarez-Pellitero2002; Fioravanti et al., Reference Fioravanti, Caffara, Florio, Gustinelli and Marcer2006), and Kudoa iwatai Egusa and Shiomitsu, 1983 in the muscle and other organs (Diamant et al., Reference Diamant, Ucko, Paperna, Colorni and Lipshitz2005). Additionally, an unnamed Henneguya species infecting the bulbus arteriosus of D. labrax has been reported from St. Gilla lagoon in Sardinia (Italy) (Culurgioni et al., Reference Culurgioni, Murtas, Cannella and Figus2010). A novel Henneguya species is described from the bulbus arteriosus of European seabass cultured in a southern Portuguese fish farm, based on a combination of morphological, biological and molecular data. This is the first description of a Henneguya species from Portugal, despite numerous myxobolids having been reported from Portuguese freshwater and estuarine environments (Saraiva and Chubb, Reference Saraiva and Chubb1989; Cruz et al., Reference Cruz, Ferreira and Saraiva1998, Reference Cruz, Saraiva and Ferreira2000; Molnár et al., Reference Molnár, Eszterbauer, Marton, Székely and Eiras2012; Rocha et al., Reference Rocha, Azevedo, Alves, Antunes and Casal2019a, Reference Rocha, Casal, Alves, Antunes, Rodrigues and Azevedo2019b; Guimarães et al., Reference Guimarães, Casal, Alves, Araújo and Rocha2023). This species was discovered in the course of research projects on Myxozoa and Coccidia parasites in farmed fish species.
Materials and methods
Sampling of European seabass
Ninety-six specimens of European seabass (D. labrax) were obtained from a fish farm located in the Alvor estuary, Atlantic coast (37°08′N, 8°37′W), Portimão, in Algarve, southern Portugal, from May 2019 to July 2021. Fresh specimens were weighted (g) and measured (cm) prior to being dissected for performing a myxosporean survey of internal organs. Due to mandatory confinements related to the COVID-19 pandemic, fish samplings could not be performed periodically, rendering it impossible to determine the seasonal prevalence of infections.
Myxosporean survey and light microscopy analyses
Several organs, including the heart, spleen, gills, liver, kidneys, gallbladder, intestine and urinary bladder were examined macroscopically using a Zeiss Stemi 2000-C binocular microscope (Grupo Taper, Sintra, Portugal) to detect structural changes or the presence of plasmodia. Microscopic examination of fresh tissue smears was performed using a Zeiss Axiophot microscope (Grupo Taper). Fresh mature myxospores were measured and photographed using an optical microscope equipped with a Zeiss Axiocam digital camera Icc3 and Zeiss Axiovision 4.6.3 software (Grupo Taper). Measurements were performed as recommended by Lom and Arthur (Reference Lom and Arthur1989) for Henneguya myxospores and are presented as mean ± s.d. (range), followed by the number of myxospores measured for each character.
DNA extraction, amplification and sequencing
DNA from ethanol-preserved myxospores of 2 isolates from 2 individual hosts (R1H and R2H) was separately extracted using the commercial kit GenElute™ Mammalian Genomic DNA Miniprep Kit (Sigma-Aldrich, Merck Life Science S.L.U., Algés, Portugal), following the manufacturer's instructions. Amplification of the parasite's small subunit ribosomal RNA (SSU rDNA) gene was achieved through the polymerase chain reaction (PCR) using a combination of universal eukaryotic primers and myxozoan-specific primers (Table 1). PCRs were performed in 25 μL reactions containing 2.5 mm of MgCl2, 1×Taq DNA polymerase reaction buffer, 1.25 U of Taq polymerase (NZYTaq II DNA polymerase, NZYTech, Lisbon, Portugal), 0.2 mm deoxyribonucleotide triphosphates (dNTPs) and 0.2 μm of each primer (NZYTech). Amplifications were performed in a Bio-Rad-MJ Mini Gradient Thermal Cycler with initial denaturation at 95°C for 3 min, followed by 35 cycles of denaturation at 94°C for 35 s, annealing at 55°C for 35 s, extension at 72°C for 45 s and a final extension at 72°C for 7 min. Five microliters of the PCR products were screened in a 1% agarose gel in TAE buffer (NZYTech) to confirm amplification. PCR products were purified, and sequenced using the same PCR primers (Table 1) by STAB VIDA (Caparica, Portugal) in a BigDye Terminator v3.1 from the Applied Biosystems Kit and run on an ABI3730XL DNA analyser (Applied Biosystems, STAB VIDA Lda., Caparica, Portugal).
Table 1. Primers used to amplify and sequencing the SSU rDNA of Henneguya myxospores found infecting the bulbus arteriosus of Dicentrarchus labrax

Phylogenetic analysis
The partial sequences obtained from the PCR products of the 2 isolates were separately assembled using ClustalW in MEGA X software (Nei and Kumar, Reference Nei and Kumar2000; Kumar et al., Reference Kumar, Stecher, Li, Knyaz and Tamura2018). The newly obtained SSU rDNA sequences were then subjected to BLASTn search (NCBI) to identify similar sequences deposited in the GenBank database. For phylogenetic analyses, 67 SSU rDNA sequences from myxobolids were retrieved, encompassing all sequences with higher than 85% similarity to the isolates under study. This included all other heart infecting Henneguya spp. that have been sequenced to date, namely Henneguya aegea Katharios, Varvarigos, Keklikoglou, Ruetten, Sojan, Akter, Cascarano, Tsertou and Kokkari, Reference Katharios, Varvarigos, Keklikoglou, Ruetten, Sojan, Akter, Cascarano, Tsertou and Kokkari2020, Henneguya akule Work, Takata, Whipps and Kent, Reference Work, Takata, Whipps and Kent2008, Henneguya cynoscioni Dyková, Buron, Roumillat and Fiala, Reference Dyková, Buron, Roumillat and Fiala2011, Henneguya lateolabracis Yokoyama, Kawakami, Yasuda and Tanaka, Reference Yokoyama, Kawakami, Yasuda and Tanaka2003, Henneguya mauritaniensis Khlifa, Miller, Adlard, Faye and Sasal, Reference Khlifa, Miller, Adlard, Faye and Sasal2012 and Henneguya pagri Yokoyama, Itoh and Tanaka, Reference Yokoyama, Itoh and Tanaka2005. The only exception was the sequence of Henneguya zikaweiensis Sikama, Reference Sikama1938 (KR020026), which infects the heart of the freshwater cyprinid Carassius auratus (Linnaeus, 1758) but shares only 75.1% similarity with the isolates under study. The SSU rDNA sequence with GenBank accession no. OR699454 was selected as representative of the several sequences (OR699454, OR699456, OR699457, OR699458, OR699460 and OR699463) deposited in this database, belonging to an unnamed Henneguya sp. from Chirodactylus variegatus (Valenciennes, 1833) in Peru, sharing 91.4–91.6% similarity with the isolates under study. Myxidium lieberkuehni Bütschli, 1882 (X76638) and Zschokkella auratis Rocha, Casal, Rangel, Severino, Castro, Azevedo and Santos, 2013 (KC849425) were used as outgroup. Alignments were performed using MAFFT version 7, available online. Phylogenetic trees were obtained using both maximum-likelihood (ML) and Bayesian inference (BI) analyses. ML analysis was performed using the PhyML 3.0 program (Dereeper et al., Reference Dereeper, Guignon, Blanc, Audic, Buffet, Chevenet, Dufayard, Guindon, Lefort, Lescot, Claverie and Gascuel2008; Guindon et al., Reference Guindon, Dufayard, Lefort, Anisimova, Hordijk and Gascuel2010) available at https://www.phylogeny.fr/. The general time reversible (GTR) model was selected based on the lowest score of the Bayesian information criterion and corrected Akaike information criterion, and bootstrap confidence values were calculated from 1000 replicates. BI analysis was performed in MrBayes v.3.2.6 (Ronquist and Huelsenbeck, Reference Ronquist and Huelsenbeck2003), with the GTR model with gamma-shaped rate variations across sites (Invgamma) (GTR + I + G) model evolution. Posterior probabilities were calculated using the Markov chain Monte Carlo method, with 4 chains running simultaneously for 1 million generations, burn-in set at 25% and trees sampled every 500 generations.
Results
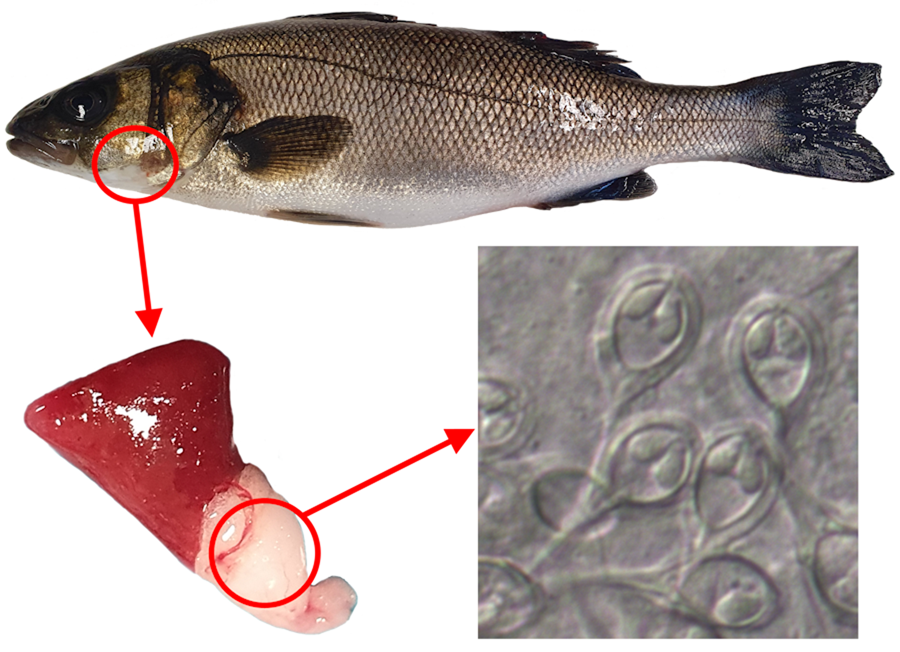
Ninety-six specimens of D. labrax were examined. The sample consisted of 35 females, 29 males and 32 fish of undetermined sex, weighing 145.0 ± 72.5 (38.0–340.2) g, and measuring 22.5 ± 3.8 (14.7–30.6) cm in length. Thirteen specimens (13.5%), 4 females, 3 males and 6 of undetermined sex, presented myxosporean infection in both the heart (bulbus arteriosus) and spleen. Infected fish weighed 149.2 ± 49.6 (74.0–218.4) g and measured 23.2 ± 2.6 (19.0–26.7) cm in length. Numerous immature and mature myxospores could be observed within whitish irregularly shaped plasmodia, around 1 mm in size, located in the bulbus arteriosus (Fig. 1), while only loose mature myxospores were detected in fresh smears of the spleen. Morphological examination of myxospores and molecular analyses of the SSU rDNA gene identified the parasite as a new member of the genus Henneguya, herein described as Henneguya cardii n. sp.

Figure 1. Irregular white plasmodia of Henneguya cardii n. sp. in the bulbus arteriosus (circle) of Dicentrarchus labrax.
Taxonomic summary
Myxobolidae Thélohan, 1892
Henneguya Thélohan, 1892
Henneguya cardii n. sp.
Type-host: European seabass Dicentrarchus labrax (Linnaeus, 1758) (Moronidae).
Type-locality: Alvor estuary, Atlantic coast (37° 08′ N, 8° 37′ W), Portimão, Algarve, Portugal.
Site of infection: Bulbus arteriosus.
Prevalence: 13.5% (13 out of 96).
Type-material: Series of phototypes of the hapantotype, deposited together with a representative DNA sample in the Type Material Collection of the Laboratory of Animal Pathology, Interdisciplinary Centre of Marine and Environmental Research, Porto, Portugal, reference CIIMAR.2024.90.
Representative DNA sequences: Two SSU rDNA sequences (GenBank accession nos. ON119140 and ON119141).
Zoobank registration: LSID urn:lsid:zoobank.org:act:3E4A797A-D8D1-4119-85F9-E0C2B0ACE859.
Etymology: The specific name ‘cardii’ refers to the heart as the site of infection.
Myxospore description
Myxospores broadly obovoid in frontal view and ellipsoidal in lateral view, with 2 equal caudal appendages forking slightly dislocated from the base. Suture line prominent. Two polar capsules, ovoid and symmetric, each comprising a polar tubule with 3–4 coils (Figs 2 and 3). Mature myxospores measuring 10.2 ± 0.6 (8.8–11.6) (n = 100) μm in length, 8.0 ± 0.7 (5.3–8.8) (n = 50) μm in width and 5.6 ± 0.2 (5.1–6.4) (n = 50) μm in thickness. Caudal appendages 36.6 ± 3.2 (27.4–42.9) (n = 28) μm long. Total spore length 47.6 ± 3.2 (41.2–53.2) (n = 20) μm. Polar capsules measuring 4.0 ± 0.2 (3.4–4.6) (n = 66) μm in length and 2.2 ± 0.1 (1.9–2.6) (n = 66) μm in width.

Figure 2. Mature myxospores of H. cardii n. sp. from the bulbus arteriosus of D. labrax: (A) myxospores from a ruptured plasmodium, (B) myxospore in valvular view and (C) myxospores in sutural view.

Figure 3. Line drawings of H. cardii n. sp. myxospores: (A) valvular view and (B) sutural view.
Remarks
Myxospore morphometric variation was not detected between different isolates. Morphological comparison with other heart infecting Henneguya species revealed similarities to Henneguya otolithi Ganapati, Reference Ganapati1941 and H. lateolabracis (see Table 2). However, H. cardii n. sp. differs from H. otolithi by being slightly shorter (8.8–11.6 vs 10.0–12.0 μm), thicker (5.1–6.4 vs 4.0–5.0 μm) and lacking a thickening running the myxospore body transversally (Ganapati, Reference Ganapati1941). Despite some measures of H. cardii n. sp. overlapping with those of H. lateolabracis (Yokoyama et al., Reference Yokoyama, Kawakami, Yasuda and Tanaka2003), the SSU rDNA sequence of these species share only 87.4% similarity. Although H. cardii n. sp. shares similar myxospore shape with the unnamed Henneguya sp. previously reported from the heart of D. labrax in Sardinia (Culurgioni et al., Reference Culurgioni, Murtas, Cannella and Figus2010), morphometric comparison to this species is impossible due to the lack of measurements in the species report.
Table 2. Myxospore morphometry of Henneguya species infecting the heart, including the newly described Henneguya cardii n. sp.

Measurements are given in micrometres. Means are provided with the standard deviation and the range in parentheses, when available.
CAL, caudal appendages length; PCL, polar capsule length; PCS, polar capsule size (=, polar capsules of equal size); PCW, polar capsule width; PTC, polar tubule coils; SBL, myxospore body length; SBT, myxospore body thickness; SBW, myxospore body width; TSL, total myxospore length; –, absence of data.
Overall, the species morphometrically more similar to H. cardii n. sp. are Henneguya clini Reed, Basson, Van As and Dyková, Reference Reed, Basson, Van As and Dyková2007, Henneguya joalensis Kpatcha, Faye, Diebakate, Fall and Toguebaye, Reference Kpatcha, Faye, Diebakate, Fall and Toguebaye1997, Henneguya lutjani Kpatcha, Faye, Diebakate, Fall and Toguebaye, Reference Kpatcha, Faye, Diebakate, Fall and Toguebaye1997 and Henneguya tegidiensis Nicholas and Jones, Reference Nicholas and Jones1959. Henneguya cardii n. sp. and H. clini share nearly identical myxospore body length (10.2 vs 10.0 μm), body width (8.0 vs 7.9 μm), caudal appendage length (36.6 vs 34.0 μm), polar capsules size (4.0 × 2.2 vs 4.0 × 2.2 μm) and total myxospore length (47.6 vs 44.6 μm). However, these species differ in site of infection (heart vs gills), number of polar tubule coils (3–4 vs 4–5), host taxa (Moronidae vs Clinidae) and geographic distribution (limited to the North Atlantic vs limited to South Africa) (Reed et al., Reference Reed, Basson, Van As and Dyková2007). Henneguya cardii n. sp. differs from H. joalensis by having a slightly larger myxospore body (10.2 × 8.0 vs 8.9 × 6.3 μm) and smaller polar capsules (4.0 × 2.2 vs 4.7 × 2.5 μm) (see Kpatcha et al., Reference Kpatcha, Faye, Diebakate, Fall and Toguebaye1997); from H. lutjani by having smaller myxospore body length (10.2 vs 11.6 μm), larger myxospore body width (8.0 vs 7.2 μm) and divergent polar capsules (4.0 × 2.2 vs 3.8 × 2.9 μm) (see Kpatcha et al., Reference Kpatcha, Faye, Diebakate, Fall and Toguebaye1997) and from H. tegidiensis by being less thick (5.6 vs 6.7 μm) and having a distinct host environment (marine vs fresh water) (see Nicholas and Jones, Reference Nicholas and Jones1959).
Molecular analysis
Isolates R1H (ON119140) and R2H (ON119141) generated SSU rDNA sequences with 2016 and 2025 bp, respectively. These were 100% identical between each other, and BLASTn search did not retrieve highly similar SSU rDNA sequences from the GenBank database. The most similar sequences belonged to an unnamed Henneguya sp. (OR699454, OR699456, OR699457, OR699458, OR699460 and OR699463) from the Peruvian morwong C. variegatus (Valenciennes, 1833) (91.4–91.6%), Henneguya lagunensis de Azevedo, Negrelli, de Oliveira, Abdallah, Camara, Matos and Vieira, 2021 (MT676010) from Brazilian mojarra Eugerres brasilianus (Cuvier, 1830) (90.9%), H. pagri (AB183748) from red seabream Pagrus major (Temminck and Schlegel, 1843) (90.8%), H. mauritaniensis (JQ687060) from bluespotted seabream Pagrus caeruleostictus (Valenciennes, 1830) (90.7%) and H. cynoscioni (JN017203) from spotted weakfish Cynoscion nebulosus (Cuvier, 1830) (90.4%).
ML and BI analyses yielded similar topologies (Fig. 4). The isolates of H. cardii n. sp. clustered within a well-supported clade comprising other Myxobolus and Henneguya spp. that have also been reported from marine environments, and that mostly infect fish belonging to Eupercaria incertae sedis, but also Carangiformes, Blenniiformes, Acropomatiformes and Centrarchiformes. The sequences of other Henneguya spp. that have been reported from the heart were also grouped within this clade, namely H. aegea, H. akule, H. cynoscioni, H. lateolabracis, H. mauritaniensis and H. pagri. Another marine clade of myxobolids was retrieved and included solely the sequences of Myxobolus spp. reported from Mugiliformes. Overall, a host-associated phylogenetic pattern was retrieved in our analyses, with species that infect Siluriformes, Characiformes, Perciformes, Centrarchiformes, Mugiliformes, Carangiformes and Eupercaria incertae sedis mostly forming well-supported clades. Some entropy was retrieved within the clade comprising H. cardii n. sp. and other marine Myxobolus/Henneguya, but this is most likely the result of low inner nodal supports.

Figure 4. Tree topology resulting from the ML analysis of 65 SSU rDNA sequences from myxobolids, including H. cardii n. sp. (sequences within grey box), and all other marine heart-infecting Henneguya spp. (sequences in bold). Numbers at the nodes are ML bootstrap values/BI posterior probabilities; dashes represent a different branching of the BI tree or a bootstrap support value under 50. Species reported from brackish/marine environments are indicated by black squares (■). The remaining species were reported from freshwater environments. Information on host taxonomy is given on the right.
Discussion
Currently, the description and taxonomic classification of myxosporeans rely on the combined analysis of several criteria, which include myxospore morphology, host and tissue specificity and molecular data (Atkinson et al., Reference Atkinson, Bartošová-Sojková, Whipps, Bartholomew, Okamura, Gruhl and Bartholomew2015). This holistic approach overcomes the artificiality of traditional morphology-based taxonomy, allowing reliable differentiation among congeners that share high morphological similarity. Nonetheless, the great majority of Henneguya species lack DNA data, making it inevitable to resort to myxospore morphology when identifying species that belong to this genus. The description of H. cardii n. sp. in this study was based on a comprehensive analysis of morphological, biological and molecular data. The species molecularly most similar to H. cardii n. sp. were an unnamed Henneguya sp. from the Peruvian morwong C. variegatus, H. lagunensis, H. pagri, H. mauritaniensis and H. cynoscioni, ranging between 90.4 and 91.5% similarity. Thus, the smallest divergence to H. cardii n. sp. was 8.5%, meaning that H. cardii n. sp. is clearly distinct from GenBank records. All congeners without molecular data could be differentiated from H. cardii n. sp. based on morphological aspects and host species. An unnamed Henneguya sp. previously reported from the heart of D. labrax in Sardinia, Mediterranean Sea (Culurgioni et al., Reference Culurgioni, Murtas, Cannella and Figus2010), was observed to share some morphological similarity to H. cardii n. sp. Nonetheless, a formal comparison could not be performed due to the lack of myxospore measurements or molecular information from the Italian species report.
Henneguya spp. are typically parasites of fish gills but have also been described from several other organs, including the heart. The concomitant occurrence of Henneguya in the heart and other organs has been suggested to be the result of myxospore spreading through the bloodstream, as with Henneguya visceralis, which develops in the kidneys, disseminating via blood to the heart, liver, spleen, gallbladder, and other organs and tissues (Jakowska and Nigrelli, Reference Jakowska and Nigrelli1953). On the contrary, myxospores of H. aegea develop in the heart prior to being disseminated to other organs through the bloodstream (Katharios et al., Reference Katharios, Varvarigos, Keklikoglou, Ruetten, Sojan, Akter, Cascarano, Tsertou and Kokkari2020). The same was observed in H. cardii n. sp., which sporogonic development occurs in the heart, with mature myxospores being seemingly transported to the spleen, where they could be observed isolated.
The phylogenetic analyses performed in this study revealed H. cardii n. sp. positioned within a well-supported lineage of myxobolids that infect marine hosts, and that is predominantly comprised by Henneguya spp., including few Myxobolus representatives. The caudal appendages of Henneguya have been suggested to optimize the dispersion capacity of myxospores in freshwater habitats or over longer distances (Liu et al., Reference Liu, Lövy, Gu and Fiala2019). Thus, the predominance of the Henneguya morphotype in the marine lineage of myxobolids strengthens the contention that character evolution is a complex and dynamic process that cannot be fully comprehended based on a single criterion, given that it most likely involves several host- and environment-related factors. For instance, mugiliform-infecting myxobolids mostly belong to the genus Myxobolus, whose morphotype has been hypothesized to optimize transmission to the deposit-feeding oligochaetes that host their actinospore development in marine habitats (Rocha et al., Reference Rocha, Rangel, Casal, Azevedo, Rodrigues and Santos2020). Discover of additional Myxobolidae life cycles would allow a better understanding of myxospore morphological character evolution.
In agreement with previously published cladograms of myxobolids (Carriero et al., Reference Carriero, Adriano, Silva, Ceccarelli and Maia2013; Liu et al., Reference Liu, Lövy, Gu and Fiala2019), our phylogenetic analyses retrieved host-associated clustering. A tendency of gill and muscle-infecting myxobolids to cluster according to the site of infection has also been shown in the literature (Eszterbauer, Reference Eszterbauer2004; Molnár and Eszterbauer, Reference Molnár, Eszterbauer, Okamura, Gruhl and Bartholomew2015) and suggests tissue tropism as a potential minor evolutionary driver for this myxosporean group. Accordingly, the tree topology obtained in this study supports a close relationship between marine heart-infecting Henneguya species. The overall low support of the marine lineage inner nodes suggests that a hidden diversity of marine myxobolids is yet to be explored that could ultimately provide better resolution of large- and fine-scale factors influencing evolutionary patterns.
Although clinical signs of infection by H. cardii n. sp. were not observed in this study, a histopathological assessment is required to evaluate the potential pathological impact that this myxosporean might have in the production of European seabass.
Data availability statement
All data supporting the findings of this study are available within the paper. Sequence data are available on the NCBI GenBank database.
Acknowledgements
We thank Dr David Gibson for his support with the choice of the specific name of the new species.
Author contributions
All authors contributed to the study conception and design, and for the financial funding. Fish samples were provided by R. S. Material preparation, data collection and analysis were performed by L. F. R. and S. R. The first draft of the manuscript was written by L. F. R. and all authors commented on previous versions of the manuscript. All authors read and approved the final manuscript.
Financial support
This research was supported by national funds through FCT – Foundation for Science and Technology (L. F. R. and M. J. S., grant numbers UIDB/04423/2020 and UIDP/04423/2020), [L. F. R., employment contract CEECIND/03501/2017/CP1420/CT0010 (https://doi.org/10.54499/CEECIND/03501/2017/CP1420/CT0010)], [S. R., project PTDC/BIA-BMA/6363/2020 (http://doi.org/10.54499/PTDC/BIA-BMA/6363/2020) and employment contract 2022.06670.CEECIND/CP1735/CT0007 (https://doi.org/10.54499/2022.06670.CEECIND/CP1735/CT0007)].
Competing interests
None.
Ethical standards
All applicable international, national and/or institutional guidelines for the care and use of animals were followed.