Introduction
Three-dimensional (3D) X-ray computed tomography (CT) of in situ dynamic processes provides internal snapshot images as a function of time. Tomograms are mathematically reconstructed from a series of radiographs taken in rapid succession as the specimen is rotated in small angular increments. In addition to spatial resolution, temporal resolution is important. Temporal resolution indicates how close together in time two distinct tomograms can be acquired. Tomograms taken in rapid succession allow detailed analyses of internal processes that cannot be obtained by other means. This article describes the state-of-the-art for such measurements acquired using synchrotron radiation as the X-ray source.
Lab-based CT
Laboratory-based micro-scale X-ray CT, first developed in 1982 by Elliot and Dover [Reference Elliott and Dover1], is often used to image (in 3D) samples in a static state. With temporal resolutions of ~1 h to 36 h, this technique may be used for the imaging of dynamic processes that occur over long periods of time, similar to time-lapse photography. A recent example of time-lapse CT was published by Lowe et al., in which the authors imaged a living butterfly chrysalis at different stages of metamor-phosis [Reference Lowe2]. For fast dynamic processes that are not easily controlled, a “post mortem” imaging approach is typically taken, in which X-ray images (that is, tomograms) are acquired before and after the dynamic process [Reference Gray3, Reference Yufit4]. For processes that can be easily controlled, a “quasi-static” (also referred to as “interrupted in situ”) approach is taken [Reference Patterson5, Reference Padilla6]. With this approach, a sample is incrementally exposed to the dynamic process, such as tensile or compressive strains, with tomograms acquired in between each interrupted step; however, some information is lost when the dynamic process is paused (for example, crack propagation) while the tomogram is acquired. Often, either the dynamic process may not be paused, or some information is lost during the pause and therefore the interrupted in situ modality is not possible. For example, the interrupted in situ compression of soft materials (for example, polymer foams) has been investigated with laboratory-based micro-scale CT, by taking a full tomogram at incremental compression steps [Reference Patterson5]. A simple observation of the stress-strain curve (see Figure 7A in reference Reference Patterson5) of a silicone foam shows that stress relaxation of the polymer foam occurs as a result of the applied uniaxial compression. Therefore, in between compression steps, a pause of typically 5 to 15 minutes is needed before the tomogram is acquired. Without this pause, residual motion within the sample leads to blurring of the tomogram and a complete loss of stress relaxation information.
Synchrotron-based CT
For adequate imaging of systems undergoing fast dynamic processes, faster imaging is needed. This requires a high photon flux of the X-ray source, high quantum efficiency of the scintillator (that is, material that converts X-ray photons to visible light photons), and a fast-frame-rate imaging detector. Synchrotron-based X-ray CT, first hypothesized by Grodzins in 1983 [Reference Grodzins7, Reference Grodzins8] and first realized by Thompson et al. in 1984 [Reference Thompson9], offers the ideal solution for this problem. Third-generation synchrotron sources [Reference Bilderback10], such as Argonne National Laboratory’s Advanced Photon Source, can offer X-ray photon fluxes ~1012 to 1014 photons s-1, which is 3 to 5 orders of magnitude brighter than what is available in the laboratory. With the availability of high-speed optical cameras, 4D (the fourth dimension being time) synchrotron-based CT is readily achievable.
Many experiments involving synchrotron-based CT are similar to the lab-based post mortem [Reference Babout11] or interrupted in situ [Reference Bari12, Reference Caty13] experiments. However, recent studies have focused on fast imaging, with temporal resolutions on the order of sub-second to tens of minutes. Recent examples include the investigation of insect mechanics [Reference Walker14, Reference dos Santos Rolo15], simultaneous heating and continuous compression of an aluminum alloy [Reference Buffière16], continuous tensile testing of matrix composites coupled with acoustic emission characterization [Reference Maire17], investigation of creep damage in a brass alloy [Reference Pyzalla18], the solidification of Al-Cu alloys [Reference Clarke19], and investigations of fatigue crack growth in SiC particle-reinforced aluminum alloy matrix composites [Reference Hruby20].
A current “hot topic” in engineering is additive manufacturing (AM) (also referred to as 3D printing or rapid prototyp-ing), which is the building of 3D structures layer-by-layer with minimal material waste, as opposed to traditional casting or machining methods. For a recent review on the history and current developments of AM, see reference [Reference Gross21]. With AM, unique structures are possible, especially with foams and lattice-structured materials that do not rely on stochastic foaming or pore-templating agents in order to produce voids [Reference Compton and Lewis22]. Foam structures using AM may be created through direct engineering.
Herein, we describe synchrotron-based CT imaging of two advanced cellular materials undergoing dynamic, continuous compression (10-2 s-1 strain rate), with a tomographic temporal resolution of 1 s and spatial resolution of ~15 μm.
Materials and Methods
Foam and microlattice
The first sample described is a 3D printed foam, which was fabricated using an Objet500 Connex 3D printer (Stratasys Ltd., Eden Prairie, MN). The printed material was a rubber-like elastomer (TangoBlack-FullCure 970). This foam exhibits parallel tubular pores in the horizontal direction. The pores exhibit a diameter of 0.75 mm and 1 mm pore spacing.
The second sample is a NiP microlattice. The microlattice was fabricated by self-propogating photopolymer waveguide prototyping, which forms a periodic polymer lattice. The metal microlattice is then fabricated from this template by coating the polymer lattice template using electroless deposition. Etching the polymer template leaves a periodic hollow tube metal microlattice. Details of microlattice fabrication can be found in references [Reference Schaedler23] and [Reference Torrents24].
Tomogram acquisition
Micro-scale X-ray CT was per-formed at the 2-BM beam line at Argonne National Laboratory’s Advanced Photon Source. Details of the tomography setup have been described elsewhere [Reference De Carlo25, Reference De Carlo and Tieman26]. A polychromatic (pink) beam (centered at ~27 keV) was used. The high photon flux of the pink beam, as compared to a monochromatic white beam, was required for sufficient signal-to-noise in the 1 ms-radiographs. Radiographs were collected using a PCO Dimax high-speed CMOS camera equipped with a 2×Mitutoyo long working distance objective lens, which resulted in 5.8 μm isotropic voxel size in the reconstructed tomograms. Each tomogram consisted of 900 radiographs acquired over 180° sample rotation in 1 s, resulting in a radiograph every 0.2° of sample rotation at ~1 ms intervals. The camera can operate much faster than 900 frames-per-second used here. The rectangular beam geometry produced a field-of-view of 2,016 pixels horizontally (11.09 mm) by 600 pixels vertically (3.30 mm). The memory on the camera was sufficient for ~18,000 radiographs.
Laboratory-based CT operates so that the sample is rotated in discrete angular increments. Rotation motion is halted to allow for radiograph acquisition, typically on the order of seconds to minutes. However, a requirement for tomogram acquisition of fast dynamic processes is that the rotation stage must be syn-chronized with the radiograph acquisi-tion time. The short 1 ms radiograph acquisition time used in this work required the sample rotation stage to be accelerated to a constant angular velocity during data acquisition in order to be synchronized. Any varia-tion in this velocity would result in a less than ideal tomogram. Ideally, the rotation stage would rotate at a con-stant angular velocity in one direc-tion for the total duration of the data acquisition (for this study, ~100 s). However, this was not possible due to the location and length of the load stage’s power supply cord, which was located at the bottom base of the load stage. Therefore, an oscillating “washing machine” rotation was used. The details of the rotation are as follows: for each tomogram, the sample rotation stage was accelerated clockwise from 0o to 180° in 2 s to a constant angular velocity. The stage was then rotated another 180° clockwise for 1 s at this constant angular velocity, during which 900 radiographs were acquired (constituting the first tomogram). The rotation stage was then decelerated over 180° clockwise for 2 seconds, for a total of 540° rotation per data acquisition cycle. The rotation stage then stopped and reversed direction for the next tomogram, accelerating for 180° anticlockwise, maintaining a constant angular velocity for the next 180° anticlockwise during data acquisition, then decelerating for the next 180° anticlockwise. This procedure was then repeated 18 more times to acquire a total of 20 tomograms per sample.
Tomograms were recorded every fifth second. The com-pression stage was held stationary during the first tomogram then continuously compressed during the remainder of the tomographic acquisition at a strain rate of 10-2 s-1. This pro-cedure was repeated for a total of 20 tomograms, after which a flat-field background and dark-field background, each consisting of 50 radiographs, was acquired. The total duration of the 20-tomogram acquisition took place during a 100 s time span.
Compression tests
The custom loading stage (Figure 1) used for these experiments is described in reference [Reference Singh27]. Compressive loading of the sample was initiated after the first tomogram was acquired, at a strain rate of 10-2 s-1. The displacement distance and displacement rate of the loading stage was set to -2.5 mm and 18 μm s-1, respectively, for the 3D printed foam. This resulted in a total compression time of 138.8 s. For the NiP microlattice, the displacement distance and displacement rate was -1 mm and 18 μm s-1 respectively, resulting in a total compression time of 55.5 s. Therefore, compression of the NiP microlattice was halted after the 11th tomogram. The drive platen moved approximately three vertical pixels per tomogram and did not lead to image blurring. During compression, samples were not constrained by the X-ray transparent poly(methyl methacrylate) (PMMA) sleeve and were not physically attached to the compression platens. Although total data acquisition time was 100 s, a period of 15–20 minutes was required before the next sample could be imaged, to allow for data transfer from the PCO Dimax camera to the data acquisition PC. Upgrades to the camera controller will increase write and read-out speeds making it possible to increase experimental compression rates by a factor of two.

Figure 1 Photo of the loading stage and X-ray CT setup at the APS 2-BM beam line.
Data processing
Tomogram reconstruction was performed using TomoPy [Reference Gursoy28], a Python-based reconstruction program developed at APS. The data were reconstructed and saved as 16-bit slices. Filters used during reconstruction include stripe removal (for example, coif16); phase retrieval (alpha set to 1 or 5 × 10-5 for our samples), which helps to set the best contrast; normalization to maximize the grayscale; and finally a zinger (despeckle) removal [29]. Stripe removal, phase retrieval, and normalization filters are not independent of each other and require several iterations to balance for the best reconstruction. Each data set of 20 tomograms was 96 GB in size.
Reconstructed tomogram visualization and analysis was performed using Avizo Fire version 8.1 (FEI Visualization Sciences Group, Burlington, MA). Smoothing of the reconstructed tomograms of the 3D printed foam was performed using the Edge-Preserving Smoothing filter found in Avizo Fire. No post-reconstruction processing was required for the NiP microlattice tomograms because of the high signal-to-noise ratio of those tomograms. Segmenta-tion of the tomograms was performed using the Interactive Threshold module found in Avizo Fire, resulting in binary images suitable for volume rendering. Post-reconstruction image processing and visualization was performed using a Hewlett-Packard Z820 workstation equipped with 96 GB of RAM, 2 Intel Xeon quad-core processors, and a Nvidia Quadro 6000 graphics card.
Results
3D Printed Foam
Figure 2A presents slices in the XY (that is, top-down view) direction of the reconstructed tomograms of the 3D printed foam at 0%, 15%, 29%, and 45% compressive strain. As can be seen in Figures 2A and 2B, four tubular voids are present, which obviously decrease in height and width during uniaxial loading. This decrease is best visualized in the XZ (side view) direction presented in Figure 2B. While the two inner tubular voids completely collapse at the last stage of compression (45% strain), there is still void structure present in the two outer tubular voids. This is due to the lack of stress in the lateral direction caused by the limited amount of ligament material in the outer region of the voids. Figure 3 is a stress-strain curve of the 3D printed foam acquired during dynamic uniaxial compression. The curve mostly follows that of a typical elastomeric foam as described by Gibson and Ashby (see Figure 5.1a in reference [Reference Gibson and Ashby30]). The stress-strain curve exhibits a plateau region that begins at 0% compressive strain and extends to ~20% compressive strain, which corresponds to elastic buckling of the foam ligaments. From ~20% compressive strain to 45% compressive strain the stress greatly increases, indicative of densification (which is evident in the volume rendering in Figure 4). Visibly absent from the stress-strain curve is a linear region at low compressive strain, which would correspond to bending of the foam ligaments.

Figure 2 Reconstructed XY (A) and XZ (B) slices of a 3D printed foam at different stages of compression. The scale bars are in micrometers.

Figure 4 Volume rendering of the 3D printed foam at different stages of compression. The scale bar is 3,000 micrometers.
Image noise and ring artifacts in the reconstructed slices (Figure 2) made a direct grayscale-based segmentation of the foam material difficult. Image noise can arise from the performance of the scintillator, the performance of the magnifying optic, and the performance of the detector. Typically, in laboratory-based CT imaging, the acquisition time for a radiograph is increased to sufficiently reduce image noise and increase the number of X-ray photons penetrating the sample per radiograph. This effectively increases the signal-to-noise ratio, resulting in a favorable reconstructed tomogram. With fast dynamic synchrotron-based CT imaging, increasing the acquisition time by even 1 ms would introduce blurring of the sample (at this strain rate), which is considered a less desired image artifact. Ring artifacts in reconstructed tomograms arise from poor detector performance; specifically, a pixel in the detector may have different sensitivity than surrounding pixels during data acquisition, yielding a false grayscale value in the acquired radiographs. If this false grayscale value is present in the majority of radiographs, the reconstructed tomogram will contain a “ring” of higher or lower grayscale value voxels in the XY orientation (as seen in Figure 2A). Modern commercially available laboratory-based CT microscopes minimize this type of image artifact by raising and lowering the sample during sample rotation, whereas this would be difficult in fast synchrotron-based CT imaging. To minimize these image artifacts in the reconstruct tomograms, mathematical smoothing filters are often employed (for example, median smoothing filters). For this data set, an edge-preserving smoothing filter was used to smooth the data, which reserves hard edges and smooths or merges soft edges with surrounding features. Figure 5 (top) presents an XY (top-down) slice of the foam at 0% compressive strain, after applying an edge-preserving smoothing filter [Reference Weickert31] to the data set (compare to Figure 2A, top left). For this data set, the filter performs an adequate smoothing of the noise in three dimensions while keeping desired edges intact, allowing for sufficient grayscale-based segmentation of the tomogram (Figure 5, middle). The histograms of the unprocessed and filtered tomograms (Figure 5, bottom) are radically different, with an increased separation of the two grayscale distributions. By applying this filter to the tomogram, noise is greatly reduced and ring artifacts are virtually eliminated.

Figure 5 Top: The same slice as presented in Figure 2A (top left), after applying an edge-preserving smoothing filter. After the smoothing step, the tomogram was segmented (middle) for the foam material. The scale bar is in micrometers. Bottom: The grayscale histograms of the tomograms before and after applying the edge-preserving smoothing filter.
From the volume renderings at four stages of compressive strain (Figure 4), the surface roughness of the foam can be visualized. Additionally, the volume rendering allows for a better visualization of the two outer void structures at the 45% compressive strain, when compared to the reconstructed slices presented in Figure 2A. Volume renderings of the two inner tubular voids, at 0% compressive strain and 29% compressive strain, are presented in Figure 6. Segmenting the tomograms for the void structures allows for not only visualization of the void volume, but also quantification of the void height and width as a function of compression.

Figure 6 Volume rendering of the two inner voids of the 3D printed foam shown in Figure 2 at 0% compression (white, translucent) and 29% compression (blue, opaque). The scale bar is in micrometers.
NiP Microlattice
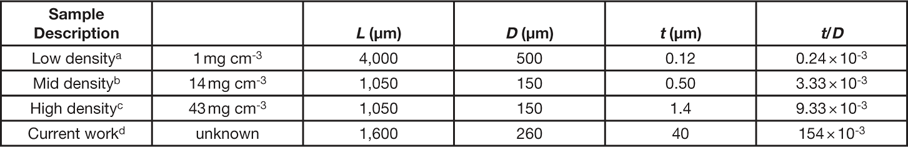
As stated in the introduction, a NiP microlattice was also imaged during uniaxial loading. This material is similar to those reported by Schaedler et al. [Reference Schaedler23]. In their report, NiP microlattices exhibited different stress responses during cyclical compressive strains, up to ~50%, which were attributed to different physical dimensions of the lattice. These physical dimensions include L, the lattice member length; D, the lattice member diameter; and t, the lattice member wall thickness (Table 1). NiP microlattices with a small wall thickness-to-diameter ratio (t/D) exhibited reversible compressive behavior and excellent recovery of their initial shape and height, whereas those with a large t/D ratio exhibited deformations typical of metallic cellular structures. For the sample examined in this work, the t/D ratio is much larger (by 2 to 3 orders of magnitude) than those that exhibit reversible compressive behavior (Table 1) because the large wall thickness was 40 µm. This resulted in a loss in plastic deformation, moving directly to brittle fracture of the microlattice, evident in the volume renderings presented in Figure 7, where a broken lattice ligament can be seen lying on the bottom of the loading stage (bottom two images of Figure 7). The stress-time curve of the NiP microlattice sample (Figure 8) exhibits high stress (~1300 kPa) at a time of 25 s, corresponding to the sixth tomogram acquired (Figure 7, top right).

Figure 7 Volume renderings of the NiP microlattice at different periods of compression. The volume renderings correspond to the 1st tomogram (top left), the 6th tomogram (top right), the 8th tomogram (bottom left), and the 13th tomogram (bottom right). The scale bar is in micrometers.

Figure 8 Stress exerted by the NiP microlattice as a function of time, acquired at the same time as the tomograms. Square symbols indicate times at which a tomogram was acquired. Black squares indicate the tomograms presented in Figure 7.
Table 1 Physical dimensions of NiP microlattices reported in reference [23] and in this work.

a See Figure 3C in reference Reference Schaedler23.
b See Figure 3A in reference Reference Schaedler23.
b See Figure 3A in reference Reference Schaedler23.
d Physical dimensions were measured from CT data.
Figure 9 presents a series of radiographs, acquired at different time intervals of the dynamic compression of a separate NiP microlattice sample. These radiographs, acquired during the 6th tomogram of the series, highlight the brittle fracturing of the NiP microlattice during compress-ion. The radiograph acquired at 7 ms (top right) exhibits significant motion blur of the NiP ligaments, due to mechanical failure of the microlattice at the ligament nodes. This failure occurs at a shorter time scale than the radiograph acquisition time, causing the blurring of the image. Also, the direction of the ligaments’ movement is perpendicular to the camera. At 396 ms, the sample has rotated 70° from the previous image, thus giving the appearance that ligament bending has not occurred, when in fact the bent ligaments are now parallel to the camera. The black arrow in this radiograph indicates a ligament that broke off of the main body of the sample at two nodes, as can be seen in the 398 ms radiograph (only 2 ms later). In the next four radiographs, the ligament falls toward the bottom of the loading stage at a calculated velocity of 2.3 km h-1 (1.4 mph). It should be noted that this particular ligament is absent in the reconstructed tomogram (not shown). This is due to the fact that the ligament is not imaged in a stationary position in each acquired radiograph that is used to reconstruct the tomogram, thus highlighting the fact that the total acquisition time for a tomogram must be significantly shorter than the desired dynamic process that is to be examined.

Figure 9 A series of radiographs of a NiP microlattice acquired during dynamic compression. The radiographs show the microlattice at different time intervals (and, consequently, at different rotation angles). The black arrow indicates a ligament that breaks off of the main body of the sample and falls to the bottom of the loading stage. The scale bar is in micrometers.
Discussion
We have demonstrated that coupling a custom loading stage to a third-generation synchrotron-based micro-scale X-ray CT beam line can result in true in situ 3D imaging of advanced cellular structures during 10-2 s-1 uniaxial compression. The compression rate was chosen so that potential blurring of the acquired tomograms was eliminated. Adequate temporal resolution dictated the spatial resolution of the CT setup. No X-ray radiation-induced damage from the high flux synchrotron X-ray source was noted on either of these samples. However, other samples (such as silicone foams) did have a noticeable yellow discoloration, indicating radiation-induced damage. Also, the PMMA sleeve used to support the upper motor did yellow in the area where the beam passed through and was replaced. Over the following weeks, it shattered within the beam path area. The largest problem with this type of data acquisition is in data handling and analysis. Large volumes of data were acquired for each sample imaged; reconstruction of a single tomogram resulted in a 4.8 GB file size, with a total of 96 GB per sample compression. Therefore, data reconstruction and data processing requires significant amounts of PC processing time and significantly depends on the hardware specifications of the processing PC. For example, the tomograms of the 3D printed foam required an edge-preserving smoothing step [Reference Weickert31], which consumed approximately five times the memory of the tomogram data set and took ~16 h to complete with our current processor.
Conclusions
The high photon flux of third-generation synchrotron sources, combined with the high-speed frame rate of commercially available camera systems, have allowed for true 4D micro-scale X-ray CT. Dynamic processes that occur on timescales within a second to tens of minutes can be adequately resolved, with an image resolution of tens of micrometers. This all depends on the sample composition, scintillator performance, synchrotron source photon flux, and frame rate of the image acquisition camera. These data are then suitable for comparison to modeling efforts, using the first tomogram as an initial input and comparing the incremental tomograms to analogous modeled data sets.
Acknowledgements
Los Alamos National Laboratory is operated by Los Alamos National Security LLC under contract number DE-AC52-06NA25396 for the US Department of Energy. Funding for this research was provided by the Enhanced Surveillance Campaign, Tom Zocco, Program Manager. The authors wish to thank Doga Gursoy with his advice in using Tomopy. This research used resources of the Advanced Photon Source, a U. S. Department of Energy (DOE) Office of Science User Facility operated for the DOE Office of Science by Argonne National Laboratory under Contract No. DE-AC02-06CH11357.