To the Editor—The determination of phenotypic antimicrobial resistance via currently available automated susceptibility systems is well established worldwide. Phenotypic testing is continuously challenged by ever-changing alterations in gene expression, genetic mutation, or new gene acquisition from another bacterium.Reference Martinez and Baquero 1 The development of antibiotic resistance increases the risk of clinical failure in infected patients, especially if such resistance is unknown to the clinical practitioner.Reference Wang, Zhang and Yao 2 The global use of automated microbiology test systems, such as MicroScan (Beckman Coulter, Brea, CA), Phoenix (Becton Dickinson Diagnostic Systems, Sparks, MD), and Vitek 2 (bioMérieux, Durham, NC), for the identification and antimicrobial susceptibility testing (AST) of bacteria has grown, but these systems have had serious reporting errors with certain organism-antibiotic combinations.Reference Sader, Fritsche and Jones 3 – Reference Kulah, Aktas, Comert, Ozlu, Akyar and Ankarali 5 We have recently identified 43 Escherichia coli isolates from 29 US hospitals that are pan-β-lactam-susceptible (ie, all cephalosporins, monobactams and carbapenems [PBL-S]) but are resistant to piperacillin-tazobactam (TZP-R), a broad-spectrum β-lactamase inhibitor.Reference Sutherland and Nicolau 6 – Reference Monogue and Nicolau 8 In this study, we assessed the accuracy of the aforementioned systems in determining the susceptibility profile of this unique phenotype.
We sent 14 unidentified clinical isolates of E. coli, 4 piperacillin-tazobactam susceptible (TZP-S)/PBL-S and 10 genotypically confirmed TZP-R/PBL-S to 3 sites for AST using MicroScan, Phoenix, and Vitek 2. To assess the accuracy of the categorical results provided by these systems (ie, susceptible, intermediate, or resistant), piperacillin-tazobactam minimum inhibitory concentrations (MICs) were determined in triplicate by broth microdilution (BMD) according to the 2016 Clinical Laboratory Standards Institute guidelines. AST data were determined via specific manufacturer and laboratory guidelines for each system. Categorical errors reported by the automated systems in relation to BMD were classified as very major (false susceptibility), major (false resistance), or minor (involving the intermediate category interpretation).Reference Rhodes, Richardson and Heraty 9
The MICs of these isolates against piperacillin-tazobactam and the interpretive classification generated by each system are reported in Table 1. Notably, none of the systems demonstrated 100% accuracy in reporting the phenotypic profile when compared with the BMD reference method. Phoenix reported 1 minor error, Vitek 2 demonstrated 2 minor errors, and MicroSca produced the most inconsistent results, with 2 very major errors and 3 minor errors.
TABLE 1 In vitro Susceptibility profile of E. coli Against Piperacillin-Tazobactam Using Broth Microdilution (BMD) and 3 Automated Susceptibility Test Systems
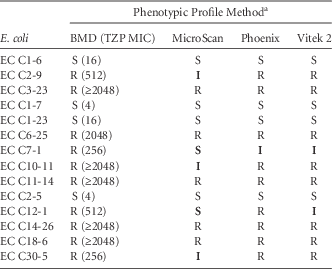
NOTE. TZP, piperacillin-tazobactam; EC, Escherichia coli; MIC, minimum inhibitory concentration (µg/mL), S, susceptible; I, intermediate; R, resistant.
a Data shown in bold are erroneous results.
These findings are relevant considering that piperacillin-tazobactam is used empirically in compromised hosts or as directed therapy for E. coli infections given the retention of high susceptibility rates compared with other available antibiotics.Reference Sutherland and Nicolau 6 , Reference Ng, Khong and Harris 10 Therefore, the detection of this TZP-R/PBL-S phenotype is vital to providing appropriate antimicrobial therapy and optimal patient care. Furthermore, the use of cascade reporting has been implemented in many hospitals to control antibiotic use, which often involves reporting the susceptibilities of broad-spectrum agents only when the organism is resistant to more narrow-spectrum agents. Therefore, cascade reporting may misrepresent the susceptibility of this organism if it is overlooked due to its susceptibility to more narrow-spectrum antimicrobial agents. Although further studies are needed to determine the clinical relevance of these TZP-R/PBL-S strains, the high use of piperacillin-tazobactam and the prevalence of E. coli infections make the recognition of this phenotype imperative because current AST systems may not accurately characterize the resistance profile. Moreover, the interpretation of AST outputs should be undertaken with caution, especially in the setting of cascade reporting.
ACKNOWLEDGMENTS
Financial support: This study was internally funded by the Center for Anti-Infective Research and Development, Hartford Hospital, Hartford, Connecticut.
Potential conflicts of interest: All authors report no conflicts of interest relevant to this article.



