Neural circuits across the brain are composed of structures that vary in size about ten million-fold. No one imaging modality can examine all the significant structures across that span. Electron microscopy (EM) excels at looking at the smallest structures, but confocal fluorescence microscopy can image the larger structures with labels to yield information about the molecular composition of the structures. Authors Ruixuan Gao, Shoh Asano, and Srigokul Upadhyayula, led by Nobel Laureate Eric Betzig and neuroscientist Edward Boyden, combined two imaging modalities that not only covered this huge range of size differences, but also identified the molecular constituents of the specimen, such as the proteins, across the range [Reference Gao1].
One of the modalities they used was expansion microscopy (ExM), which was reviewed in this column in 2015 (Microscopy Today, 23(3) (2015) 8–10). Basically this technique involves adding a water-absorbent polymer gel to preserved tissue. When transferred from a saline solution to pure water, the polymer swells, stretching the tissue approximately four-fold. The imaging relies on fluorescent tags that attach specifically to proteins in cells and tissues and also to the surrounding gel. The proteins are digested to leave the fluorescent molecules exactly in place, and the relative positions between these molecules are maintained after the swelling.
The expanded specimen was imaged by Gao et al. using lattice light-sheet microscopy (LLSM), which was also reviewed in this column (Microscopy Today, 23(1) (2015) 8–10). Briefly, this technique sweeps an ultrathin layer of light through the tissue. By using a less intense light than is required for other methods, the beam can linger on the sample while not bleaching the fluorescence or obscuring part of the image. Since it is relatively fast, LLSM illuminates a whole plane at one time rather than a single spot. It could gather enough information to image the entire brain of a fruit fly (Drosophila) in a matter of days, which would take years using EM. The imaging produces 3D image tiles, which are then assembled by computer. Using this method the team was able to map more that 40 million synapses in the brain of the fruit fly at nanometer resolution (see Figure 1). The technique yielded similar results when applied to a slice of the brain from mice.
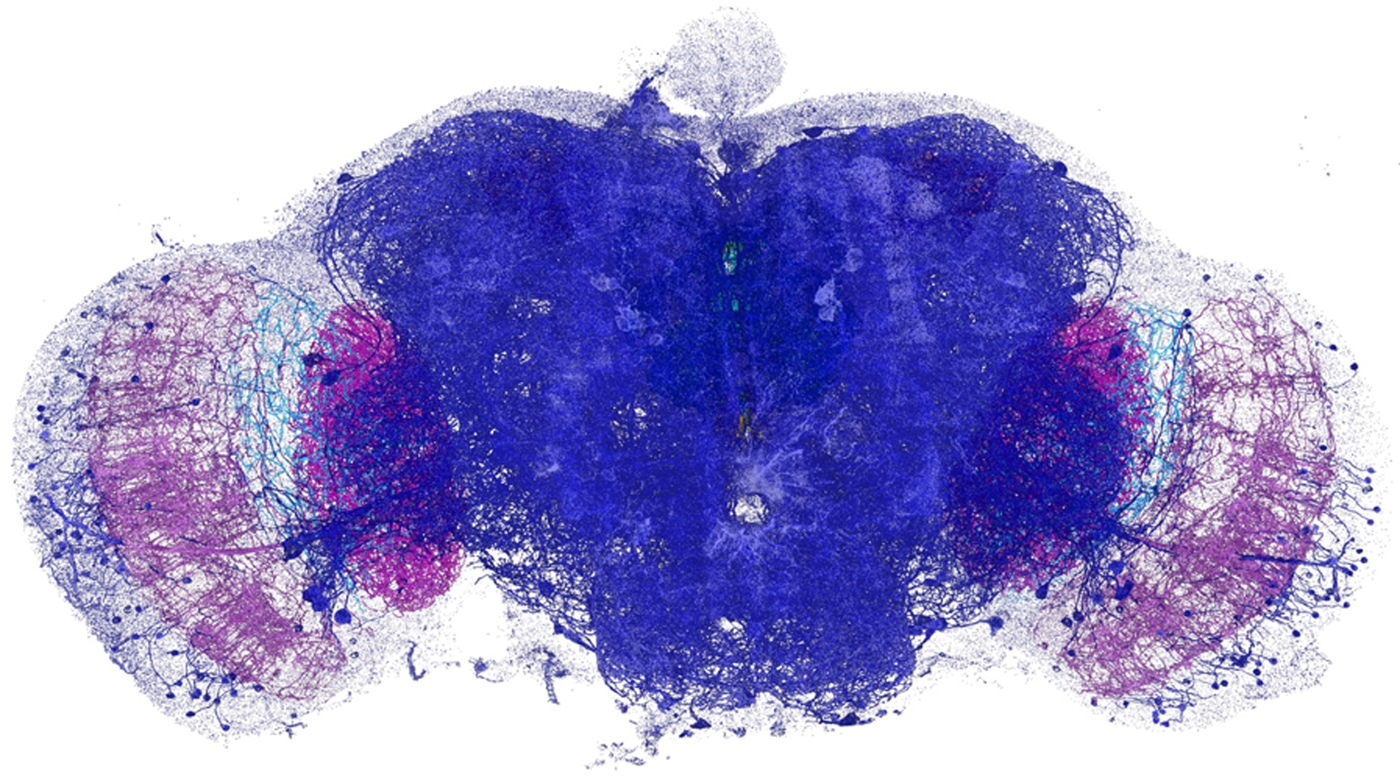
Figure 1: Dopaminergic neurons and the associated presynaptic sites across an adult fruit fly brain, color-coded by brain regions. Image width = 660 µm.
This approach can be performed at speeds that will likely enable comparative studies of neural development, circuit stereotyping, and structural correlations to neural activity or behavior. Combining ExM and LLSM fills a valuable niche between the high throughput of conventional optical imaging of neural anatomy and the ultrahigh resolution of corresponding EM imaging. Assuming the development of fully validated, brain-wide isotropic expansion at ten times or more and sufficiently dense labeling, this combination may enable comparisons throughout the brain of even densely innervated neural circuits across multiple specimens with protein-specific contrast at a resolution of 25 nanometers or better. The future is very bright for this field!




