Polymorphic markers in the brain-derived neurotrophic factor (BDNF) gene have been found to be significantly associated with bipolar disorder in two relatively large samples (Reference Neves-Pereira, Mundo and MugliaNeves-Pereira et al, 2002; Reference Sklar, Gabriel and McInnisSklar et al, 2002). However, these findings have not consistently been replicated in other studies (Reference Hong, Huo and YenHong et al, 2003; Reference Nakata, Ujike and SakaiNakata et al, 2003; Reference Kunugi, Iijima and TatsumiKunugi et al, 2004; Reference Oswald, Del-Favero and MassatOswald et al, 2004; Reference Skibinska, Hauser and CzerskiSkibinska et al, 2004; Reference Neves-Pereira, Cheung and PasdarNeves-Pereira et al, 2005). Of further interest, studies of the BDNF gene in three independent samples with child-onset mood disorder have found a significant association in each sample (Reference Geller, Badner and TillmanGeller et al, 2004; Reference StraussStrauss, 2004; Reference Strauss, Barr and GeorgeStrauss et al, 2005). The mixed findings across genetic studies of BDNF in mood disorders may be owing to the variable ascertainment of subtypes, small sizes of effect, genetic heterogeneity or other methodological variation among studies. As a first step in the process of disentangling genotype–phenotype correlations, we examined the role of BDNF gene polymorphisms in specific clinical subgroupings of bipolar disorder.
METHOD
Sample
The families were recruited in Toronto and across central Canada through newspaper advertisements and hospital clinic referrals. The procedures were approved by the research ethics board of the Centre for Addiction and Mental Health. After complete description of the study, written informed consent was obtained from each proband and family member.
The sample investigated was mostly recruited from out-patient populations, and consisted of 312 nuclear families with at least one offspring (118 males and 194 females) who had experienced at least one hypomanic or manic episode throughout life diagnosed as bipolar disorder I or II, or schizoaffective disorder, bipolar type, according to DSM–IV criteria (American Psychiatric Association, 1994). Whenever possible, siblings were included in the study. Thus, 26 siblings with bipolar disorder and 46 non-affected siblings were included. In addition, 45 first- or second-degree relatives (e.g. parents or grand-parents) were included, and 12 of these had a lifetime history of bipolar disorder. Thus, the total sample comprised 1043 people, 350 with bipolar disorder (131 males and 219 females) and 693 non-affected relatives. The distribution of bipolar disorder I and II according to DSM–IV criteria was assigned as follows: I, 200; I with rapid cycling, 27; I with seasonal patterns, 4; I with mixed episodes, 3; II, 57; II with rapid cycling, 31; II with seasonal patterns, 8. One person had a diagnosis of bipolar disorder not otherwise specified and 19 schizoaffective disorder, bipolar type. DSM–IV defines rapid cycling as the occurrence of four or more (depressive and/or manic) mood episodes within 12 months. Participants’ mean age was 35.4 years (s.d.=10.3), with a mean age at onset of 20.1 years (s.d.=7.5). Participants were mainly White and of European origin (n=332, 95%), with 12 Asians (3.4%), 3 Native Americans (0.8%), and 3 African–Americans (0.8%).
Diagnostic assessment
Details of the diagnostic assessment procedures for this sample have been published previously (Reference Carter, Mundo and ParikhCarter et al, 2003). Briefly, DSM–IV diagnoses were based on a standardised best-estimate procedure. A semistructured clinical interview (SCID–I; American Psychiatric Association, 1994) was performed by a trained research assistant who also interviewed relatives and collected information from medical records. Two experienced psychiatrists subsequently reviewed information in order to assign best-estimate consensus diagnoses. A third psychiatrist reviewed a preset percentage of all cases for quality assurance, and reviewed cases with diagnostic disagreement before a consensus diagnosis was assigned. All individuals with a diagnosis of bipolar disorder I and II were thoroughly assessed for the occurrence of rapid cycling. If a clinical subtype could not be assigned with certainty, these individuals were then excluded from our analyses.
Genotyping
The organisation of the BDNF gene is rather complex. There are at least two isoforms involving both coding and noncoding exons that are transcribed in both sense and antisense directions (Reference Liu, Walther and DrgonLiu et al, 2005). The six markers that we have analysed (Fig. 1) cover a relatively broad region of the gene. Two of the markers, the Val66Met polymorphism (NCBI SNP cluster ID: rs6265) and the GT dinucleotide repeat polymorphism (Reference Proschel, Saunders and RosesProschel et al, 1992) have been previously analysed in our sample for association with bipolar disorder (Reference Neves-Pereira, Mundo and MugliaNeves-Pereira et al, 2002). The GT(n) repeat polymorphism consists of up to ten different alleles in various populations. In our analyses, allele 1 is 174 base pairs, allele 2 is 172 base pairs, allele 3 is 170 base pairs, etc.
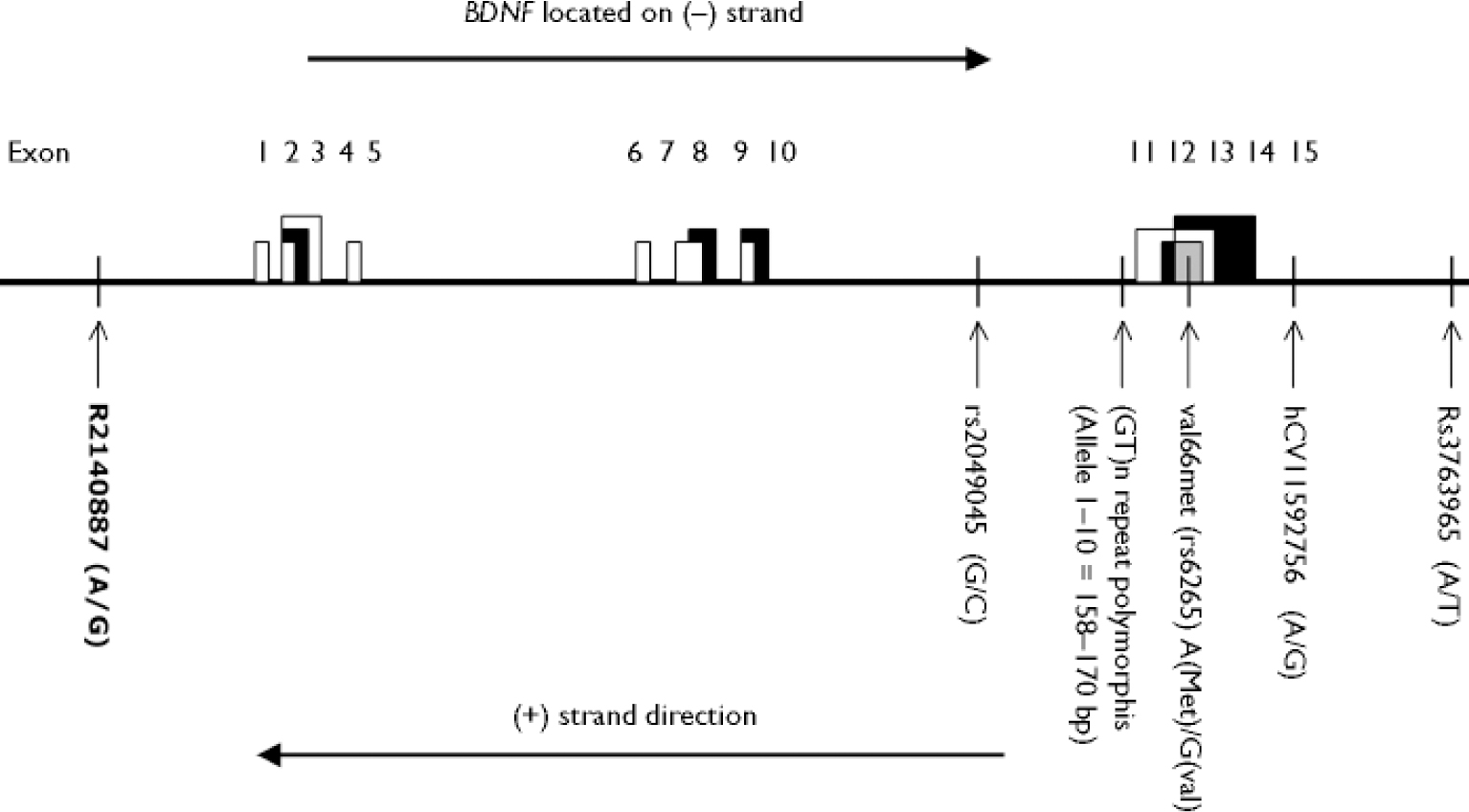
Fig. 1 Map of BDNF gene and markers used in analyses. White, grey or black coloured boxes indicate the alternate splicing. Vertical lines indicate the approximate location of BDNF gene polymorphisms. Note that the map represents a simplified model and is not to scale.
The four new markers chosen were two each in the upstream (5′) and downstream (3′) regions of the Val66Met and the GT repeat. Marker rs3763965 is located approximately 148 kb and marker hCV11592756 (Celera ID) about 7 kb upstream of the Val66Met polymorphism. Marker rs2049045 is located 13 kb downstream of the GT repeat and rs2140887 is located approximately 352 kb downstream. The reader is referred to http://www.hapmap.org for the latest haplotype interpretation of the BDNF gene.
Genotyping of the single nucleotide polymorphisms (SNPs) was performed using 5′ nuclease Taqman allelic discrimination assay on the ABI 7000 Sequence Detection System (Applied Biosystems, CA, USA). Commercially available ABI Taqman assays were used, following the manufacturers’ recommended protocol. Results were verified independently by two laboratory personnel masked to affection status.
Statistical analyses
Association tests and haplotype analyses between BDNF markers and bipolar disorder were performed using TDTPHASE (Reference DudbridgeDudbridge, 2003). Analyses were first conducted on the total sample with bipolar disorder then on the DSM–IV subtypes of bipolar disorder I and II. Finally, the total sample was divided according to rapid-cycling status: those with rapid cycling (56 nuclear families, n=180) and those without (256 nuclear families, n=863).
Linkage disequilibrium and identification of haplotype blocks within the BDNF gene were performed using HAPLOVIEW (Reference Barrett, Fry and MallerBarrett et al, 2005). The standard Lewontin D′ and correlation coefficient r2 were calculated using the expectation-maximisation algorithm implemented in HAPLOVIEW (data not shown). The GT(n) repeat polymorphism was divided into allele 3 and other alleles for the linkage disequilibrium analyses.
RESULTS
BDNF gene polymorphisms and bipolar disorder
The genotype distributions in the overall sample did not deviate from Hardy–Weinberg equilibrium at any of the six BDNF polymorphisms.
Significant associations between the total sample and the BDNF gene were found for four markers: hCV11592756, Val66Met, GT(n) repeat polymorphism, and rs2049045. Over-transmissions were observed for the A allele of hCV11592756, the Val (or G) allele of Val66Met, for allele 3 of the GT(n) repeat and for the G allele of rs2049045. Allele 3 of the GT(n) repeat was the most common, and thus was tested against the other alleles for association with bipolar disorder and bipolar disorder I and II, with and without rapid cycling, and for subsequent haplotype analyses. Findings related to the Val66Met and the GT(n) repeat polymorphism were published earlier (Reference Neves-Pereira, Mundo and MugliaNeves-Pereira et al, 2002). Two of these BDNF markers (hCV11592756 and rs2049045) were not previously known to be associated with bipolar disorder. The remaining two markers (rs3763965 and rs2140887) did not show an association with bipolar disorder (Table 2).
Table 1 Examples of individual haplotype transmission analyses of the BDNF gene markers and bipolar disorder (I and II; TDTPHASE)
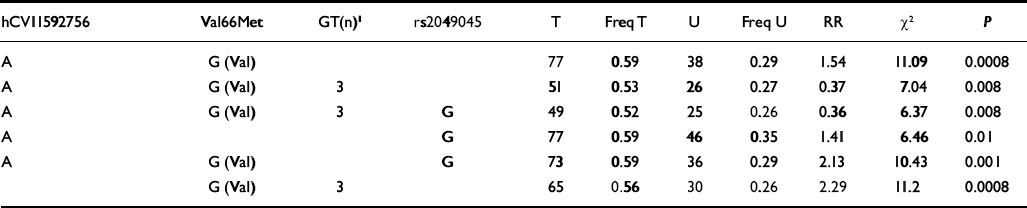
| hCVII592756 | Val66Met | GT(n)1 | rs2049045 | T | Freq T | U | Freq U | RR | X 2 | P |
|---|---|---|---|---|---|---|---|---|---|---|
| A | G (Val) | 77 | 0.59 | 38 | 0.29 | 1.54 | 11.09 | 0.0008 | ||
| A | G (Val) | 3 | 51 | 0.53 | 26 | 0.27 | 0.37 | 7.04 | 0.008 | |
| A | G (Val) | 3 | G | 49 | 0.52 | 25 | 0.26 | 0.36 | 6.37 | 0.008 |
| A | G | 77 | 0.59 | 46 | 0.35 | 1.41 | 6.46 | 0.01 | ||
| A | G (Val) | G | 73 | 0.59 | 36 | 0.29 | 2.13 | 10.43 | 0.001 | |
| G (Val) | 3 | 65 | 0.56 | 30 | 0.26 | 2.29 | 11.2 | 0.0008 |
Table 2 Association tests between BDNF markers and bipolar disorder (I and II; TDTPHASE)
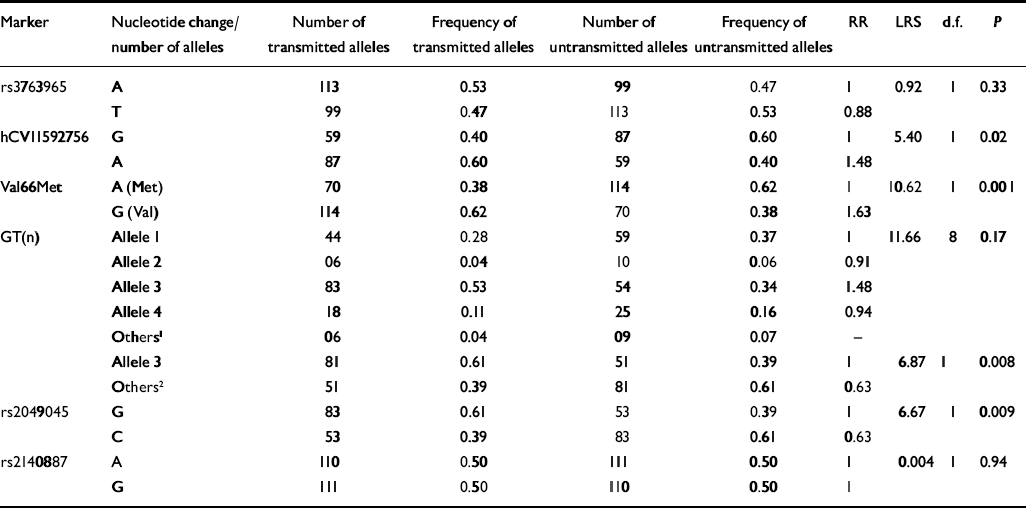
| Marker | Nucleotide change/number of alleles | Number of transmitted alleles | Frequency of transmitted alleles | Number of untransmitted alleles | Frequency of untransmitted alleles | RR | LRS | d.f. | P |
|---|---|---|---|---|---|---|---|---|---|
| rs3763965 | A | 113 | 0.53 | 99 | 0.47 | 1 | 0.92 | 1 | 0.33 |
| T | 99 | 0.47 | 113 | 0.53 | 0.88 | ||||
| hCV11592756 | G | 59 | 0.40 | 87 | 0.60 | 1 | 5.40 | 1 | 0.02 |
| A | 87 | 0.60 | 59 | 0.40 | 1.48 | ||||
| Val66Met | A (Met) | 70 | 0.38 | 114 | 0.62 | 1 | 10.62 | 1 | 0.001 |
| G (Val) | 114 | 0.62 | 70 | 0.38 | 1.63 | ||||
| GT(n) | Allele 1 | 44 | 0.28 | 59 | 0.37 | 1 | 11.66 | 8 | 0.17 |
| Allele 2 | 06 | 0.04 | 10 | 0.06 | 0.91 | ||||
| Allele 3 | 83 | 0.53 | 54 | 0.34 | 1.48 | ||||
| Allele 4 | 18 | 0.11 | 25 | 0.16 | 0.94 | ||||
| Others1 | 06 | 0.04 | 09 | 0.07 | - | ||||
| Allele 3 | 81 | 0.61 | 51 | 0.39 | 1 | 6.87 | 1 | 0.008 | |
| Others2 | 51 | 0.39 | 81 | 0.61 | 0.63 | ||||
| rs2049045 | G | 83 | 0.61 | 53 | 0.39 | 1 | 6.67 | 1 | 0.009 |
| C | 53 | 0.39 | 83 | 0.61 | 0.63 | ||||
| rs2140887 | A | 110 | 0.50 | 111 | 0.50 | 1 | 0.004 | 1 | 0.94 |
| G | 111 | 0.50 | 110 | 0.50 | 1 |
When participants with bipolar disorder were divided into two groups (bipolar disorder I and II), a significant association was found for the Val66Met polymorphism of the BDNF gene but for none of the remaining five markers. The Val (or G) allele was over-transmitted (data not shown) compared with the Met (or A) allele (bipolar disorder I: 80 v. 52 transmissions, P=0.01; bipolar disorder II: 34 v. 18 transmissions, P=0.02 respectively).
When participants with bipolar disorder were split into rapid-cycling and non-rapid-cycling subgroups, only the former showed significant associations with all four polymorphisms that had been found to be associated with the total sample (marker hCV11592756, Val66Met, GT(n), and rs2049045). Overall, the same pattern of over-transmitted alleles was observed as in the total sample. The ratio of transmitted v. untransmitted alleles was more pronounced in the rapid-cycling sampling compared with the total sample (Table 3). On the other hand, the non-rapid-cycling sample showed no significant associations with any of the six polymorphisms on the BDNF gene (Table 4).
Table 3 Association tests between BDNF markers and bipolar disorder (I and II) with rapid cycling (TDTPHASE)

| Marker | Nucleotide change/number of alleles | Number of transmitted alleles | Frequency of transmitted alleles | Number of untransmitted alleles | Frequency of untransmitted alleles | RR | LRS | d.f. | P |
|---|---|---|---|---|---|---|---|---|---|
| rs3763965 | A | 22 | 0.47 | 25 | 0.53 | 1 | 0.19 | 1 | 0.66 |
| T | 25 | 0.53 | 22 | 0.47 | 1.13 | ||||
| hCVI1592756 | G | 8 | 0.29 | 20 | 0.71 | 1 | 5.31 | 1 | 0.02 |
| A | 20 | 0.71 | 8 | 0.29 | 2.5 | ||||
| Val66Met | A (Met) | 8 | 0.24 | 25 | 0.76 | 1 | 9.19 | 1 | 0.002 |
| G (Val) | 25 | 0.76 | 8 | 0.24 | 3.12 | ||||
| GT(n) | Allele 1 | 5 | 0.15 | 18 | 0.53 | 1 | 12.67 | 5 | 0.03 |
| Allele 2 | 1 | 0.03 | 1 | 0.03 | 4.03 | ||||
| Allele 3 | 24 | 0.70 | 8 | 0.23 | 4.03 | ||||
| Allele 4 | 3 | 0.09 | 6 | 0.18 | 1.42 | ||||
| Others1 | 1 | 0.03 | 1 | 0.03 | — | ||||
| Allele 3 | 24 | 0.75 | 8 | 0.25 | 1 | 8.37 | 1 | 0.004 | |
| Others2 | 8 | 0.25 | 24 | 0.75 | 0.33 | ||||
| rs2049045 | G | 19 | 0.76 | 6 | 0.24 | 1 | 7.10 | 1 | 0.008 |
| C | 6 | 0.24 | 19 | 0.76 | 0.31 | ||||
| rs2140887 | A | 23 | 0.47 | 26 | 0.53 | 1 | 0.18 | 1 | 0.67 |
| G | 26 | 0.53 | 23 | 0.47 | 1.13 |
Table 4 Association tests between BDNF markers and bipolar disorder (I and II) without rapid cycling (TDTPHASE)
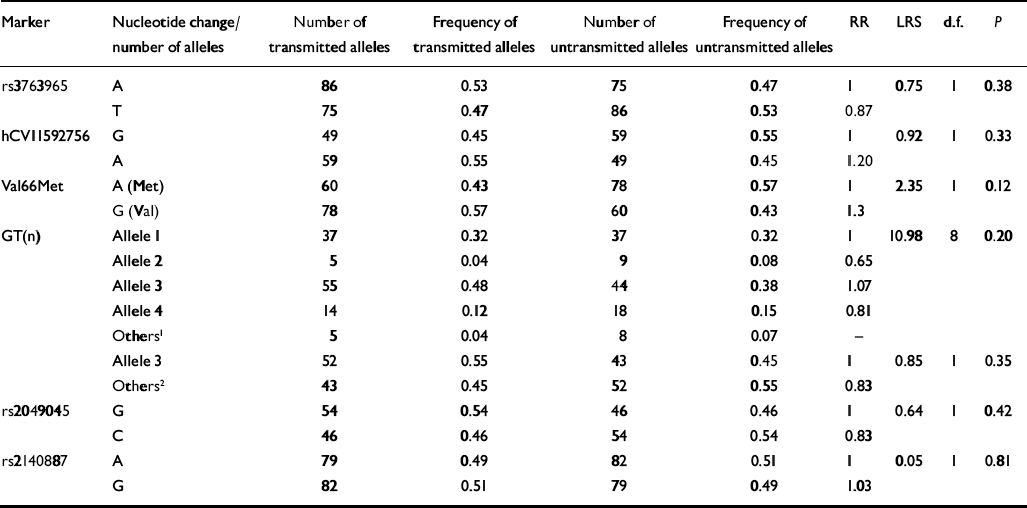
| Marker | Nucleotide change/number of alleles | Number of transmitted alleles | Frequency of transmitted alleles | Number of untransmitted alleles | Frequency of untransmitted alleles | RR | LRS | d.f. | P |
|---|---|---|---|---|---|---|---|---|---|
| rs3763965 | A | 86 | 0.53 | 75 | 0.47 | 1 | 0.75 | 1 | 0.38 |
| T | 75 | 0.47 | 86 | 0.53 | 0.87 | ||||
| hCVI1592756 | G | 49 | 0.45 | 59 | 0.55 | 1 | 0.92 | 1 | 0.33 |
| A | 59 | 0.55 | 49 | 0.45 | 1.20 | ||||
| Val66Met | A (Met) | 60 | 0.43 | 78 | 0.57 | 1 | 2.35 | 1 | 0.12 |
| G (Val) | 78 | 0.57 | 60 | 0.43 | 1.3 | ||||
| GT(n) | Allele 1 | 37 | 0.32 | 37 | 0.32 | 1 | 10.98 | 8 | 0.20 |
| Allele 2 | 5 | 0.04 | 9 | 0.08 | 0.65 | ||||
| Allele 3 | 55 | 0.48 | 44 | 0.38 | 1.07 | ||||
| Allele 4 | 14 | 0.12 | 18 | 0.15 | 0.81 | ||||
| Others1 | 5 | 0.04 | 8 | 0.07 | — | ||||
| Allele 3 | 52 | 0.55 | 43 | 0.45 | 1 | 0.85 | 1 | 0.35 | |
| Others2 | 43 | 0.45 | 52 | 0.55 | 0.83 | ||||
| rs2049045 | G | 54 | 0.54 | 46 | 0.46 | 1 | 0.64 | 1 | 0.42 |
| C | 46 | 0.46 | 54 | 0.54 | 0.83 | ||||
| rs2140887 | A | 79 | 0.49 | 82 | 0.51 | 1 | 0.05 | 1 | 0.81 |
| G | 82 | 0.51 | 79 | 0.49 | 1.03 |
Linkage disequilibrium and haplotype analyses
Analyses with HAPLOVIEW revealed that marker rs2049045 is in linkage disequilibrium with marker hCV11592756 and the Val66Met polymorphism (D′ above 0.80). A reduced linkage disequilibrium was noted for allele 3 of the GT(n) repeat polymorphism. The four markers that were individually associated with bipolar disorder proved to be in linkage disequilibrium and are part of a block within the BDNF gene. Next, we performed haplotype analyses of these four markers v. the phenotype of bipolar disorder. The A-Val-3-G haplotype proved to be significantly overtransmitted (49:25) in participants with bipolar disorder (P=0.008) (Table 1). The strongest results, however, were obtained when the Val allele was combined with either marker hCV11592756 or allele 3 of the GT(n) repeat polymorphism (P=0.0008, Table 1). We then performed haplotype analyses in the rapid-cycling sample. Although the combination of the Val allele with either marker hCV11592756 or allele 3 of the GT(n) repeat polymorphism yielded significant results, the four-marker haplotype composed of marker hCV11592756 (A allele), Val66Met (Val allele), GT(n) repeat (allele 3) and rs2049045 (G allele) yielded a nonsignificant trend (Table 5).
Table 5 Examples of individual haplotype transmission analyses of the BDNF gene marker with rapid cycling bipolar disorder (I and II; TDTPHASE)

| hCV11592756 | Val66Met | GT(n)1 | rs2049045 | T | Freq T | U | Freq U | RR | χ2 | P |
|---|---|---|---|---|---|---|---|---|---|---|
| A | G (Val) | 16 | 0.72 | 4 | 0.18 | 6.55 | 6.58 | 0.01 | ||
| A | G (Val) | 3 | 12 | 0.66 | 4 | 0.22 | 1.17e+008 | 3.76 | 0.05 | |
| A | G (Val) | 3 | G | 9 | 0.6 | 4 | 0.26 | 1.06e+008 | 1.97 | 0.16 |
| A | G | 15 | 0.75 | 5 | 0.25 | 1 | 4.81 | 0.02 | ||
| A | G (Val) | G | 14 | 0.7 | 4 | 0.2 | 5.12 | 4.91 | 0.02 | |
| G (Val) | 3 | 18 | 0.72 | 4 | 0.16 | 1.70 | 8.36 | 0.003 |
DISCUSSION
Association of four linked markers of the BDNF gene with bipolar disorder
In addition to our previously reported association between the Val66Met and the GT(n) repeat polymorphisms of the BDNF gene and bipolar disorder (Reference Neves-Pereira, Mundo and MugliaNeves-Pereira et al, 2002), we now report additional markers (hCV11592756 and rs2049045) to be associated with bipolar disorder. Further analyses indicated that these four gene variants are in linkage disequilibrium with each other, and are part of a haplotype block within the BDNF gene. In subsequent studies it may be valuable to sequence affected and unaffected individuals, to determine whether there are variants not previously known that are highly represented in affected patients and not in controls.
Potential biological impact of BDNF markers associated with bipolar disorder
The functional relevance of the GT(n) repeat polymorphism and the other SNPs (apart from Val66Met) remains unknown.
In the case of the GT(n) marker, we have categorised alleles into two groups: allele 3 v. alleles 1, 2 and 4–10. This approach, however, remains arbitrary and is different from one previous study that analysed for association between this BDNF GT(n) marker and age at onset and therapeutic response in schizophrenia, grouping alleles into longer (172–176 base pairs) v. shorter variants (166–174 base pairs) (Reference Krebs, Guillin and BourdellKrebs et al, 2000). Thus, allele grouping remains arbitrary until future studies elucidate the functional relevance of the GT(n) polymorphism.
In contrast to the other markers in the BDNF gene, relatively extensive in vitro and in vivo functional analyses have been performed for the Val66Met polymorphism. In cultured hippocampal neurons, the Val allele (vBDNF) has been shown to increase intraneuronal BDNF peptide secretion and distribution compared with the Met allele (mBDNF) (Reference Egan, Kojima and CallicottEgan et al, 2003). Neurons expressing vBDNF were shown to express BDNF in the cell body and distal processes (dendrites). In contrast, mBDNF was mainly localised in cell bodies. In humans, the Met allele was associated with poorer episodic memory, and abnormal activation of the hippocampus, as measured by functional magnetic resonance imaging (Reference Egan, Kojima and CallicottEgan et al, 2003; Reference Pezawas, Verchinski and MattayPezawas et al, 2004). Our results, and those of others that show the Val allele of Val66Met to be contributing to risk, suggest that relatively rapidly changing mood episodes might be associated with enhanced memory function (Reference Egan, Kojima and CallicottEgan et al, 2003) and increased hippocampal–frontal connectivity (Reference Pezawas, Verchinski and MattayPezawas et al, 2004). This speculation will require more investigation using cognitive testing and functional imaging techniques, preferably in individuals with rapid-cycling compared with those with other mood patterns. Alternatively, the enhanced memory may be a separate effect of the BDNF Val allele and have no biological connection to mood disorder.
In terms of the other markers across the BDNF gene, it may be that the GT(n) repeat polymorphism and BDNF variants at marker sites hCV11592756 and rs2049045 are not of functional relevance, but are associated with bipolar disorder because of relatively strong linkage disequilibrium with the functionally relevant Val66Met polymorphism. However, it is also plausible that the GT(n) and/or other markers may alter mRNA stability or processing, thus altering the amount of the BDNF peptide that is produced, increasing or decreasing the effect of the Val66Met change in the protein. The net effect on risk for bipolar disorder may thus be best captured by typing multiple markers across the gene.
BDNF: possible association with mood disorders despite inconsistent findings
Our findings obviously need to be put in the context of previous findings. As mentioned above, a number of studies failed to detect a significant association between the BDNF gene and bipolar disorder (Reference Hong, Huo and YenHong et al, 2003; Reference Nakata, Ujike and SakaiNakata et al 2003; Reference Kunugi, Iijima and TatsumiKunugi et al, 2004; Reference Oswald, Del-Favero and MassatOswald et al, 2004; Reference Skibinska, Hauser and CzerskiSkibinska et al, 2004; Reference Neves-Pereira, Cheung and PasdarNeves-Pereira et al, 2005). Further supportive evidence for association of the BDNF gene and mood disorder derives from a recent study that found an association between the Val66Met polymorphism and a prepubertal and early adolescent phenotype (Reference Geller, Badner and TillmanGeller et al, 2004). Other interesting findings derive from preliminary analyses of a study that included large samples of German descent, and revealed most significant associations between haplotypes (including the Val/Met polymorphism) in two independent samples of patients with major depression (n=465 and 312), as well as significant association in one sample with bipolar disorder (n=281) and one with schizophrenia (n=533) (Reference Cichon, Schumacher and Abou JamraCichon et al, 2004).
Inconsistent findings may be the result of general problems of molecular genetic association studies dealing with complex disorders (Reference Schulze, Hardy and McMahonSchulze et al, 2003). Some studies may be too small or under-powered to detect modest gene effects. There is also variation in terms of study design, such as ascertainment strategies and inclusion criteria. Thus far, negative findings have been observed in population-based case–control studies, whereas initial positive findings were based on family-based association studies (Reference Neves-Pereira, Mundo and MugliaNeves-Pereira et al, 2002; Reference Sklar, Gabriel and McInnisSklar et al, 2002). Indeed, it has been noted that different ascertainment strategies may introduce notable differences with respect to important clinical and demographic characteristics, particularly in samples ascertained for case–control studies v. family-based studies (Reference Schulze, Muller and KraussSchulze et al, 2001). Our sample included relatively young out-patients and patients with less severe forms of bipolar disorder (i.e. type II according to DSM–IV). In contrast, other studies have included inpatients who were likely to be older at time of inclusion and possibly affected with more chronic forms of bipolar disorder. Finally, other studies analysed samples of different ethnic backgrounds (including Chinese or Japanese), and thus the BDNF gene may represent a genetic risk factor which is more pronounced in Whites than in other ethnic groups, possibly because of local differences in and around the BDNF gene or differences in more general genetic background.
BDNF and rapid cycling
According to the concept of people with bipolar disorder forming sub-populations that share a distinct genetic liability, we analysed the well recognised subgroups, including those with rapid cycling (Reference Dunner and FieveDunner & Fieve, 1974). Rapid cycling represents a widely accepted clinical category, and is reported to occur in 5–15% of persons with bipolar disorder (American Psychiatric Association, 1994). People experiencing rapid cycling may represent a distinct subtype with respect to pharmacological response (i.e. resistance to lithium therapy) or demographic factors (i.e. female preponderance) (Reference Mackin and YoungMackin & Young, 2004). We observed that our significant association between the BDNF gene and bipolar disorder is mainly driven by the inclusion of this particular group of participants. The impact of this effect is strong, as removing the rapid-cycling participants from our analyses eliminates any positive association. This result is mirrored by another study of White people representing the largest bipolar disorder case–control sample to date, that did not detect significant associations between the Val66Met polymorphism and bipolar disorder but did detect a significant association in their subset of rapid-cycling patients (Reference Green, Raybould and MacgregorGreen et al, 2006).
Implications and limitations
The concordance of our study and that of Green et al (Reference Green, Raybould and Macgregor2006) has important implications. First, inconsistent findings in previous studies might at least partly result from not examining patients with rapid cycling. Thus, our findings should stimulate re-analysis of previous samples, selecting for patients that experienced rapid cycling. Second, it is plausible that the BDNF gene is associated with rapid mood swings or with more general mood instability. Such symptoms are not exclusively encountered in bipolar disorder, but also present in a variety of other neuropsychiatric disorders. In accordance with that hypothesis, BDNF gene polymorphisms have been associated with disorders that are characterised by mood symptoms such as schizophrenia (Reference Muglia, Vicente and VergaMuglia et al, 2003; Reference Neves-Pereira, Cheung and PasdarNeves-Pereira et al, 2005), obsessive–compulsive disorder (Reference Hall, Dhilla and CharalambousHall et al, 2003) or adult attention-deficit hyperactivity disorder (Reference Lanktree, Muglia and SquassinaLanktree et al, 2004). Third, it is also plausible that BDNF is associated with depressive symptoms without mania. Although our findings in the bipolar disorder II sample did only reveal a significant association with the Val66Met polymorphism, it remains possible that BDNF is particularly associated with bipolar disorder II. Rapid cycling is mostly encountered in patients with bipolar disorder II who have less severe manic symptoms, but who tend to have persistent treatment-resistant depression, with at least one full-blown episode of major depression. Also in line with this hypothesis are the significant findings with the BDNF gene and childhood-onset mood disorders in a sample of adults who were followed up over two decades from the childhood mood disorder, and only a minority of them became bipolar (Reference StraussStrauss, 2004). Fourth, BDNF may be associated with personality traits that may confer susceptibility to mood or depressive symptoms. One study found anxiety- and depression-related personality traits to be associated with the Val/Val genotype of the BDNF gene (Reference Lang, Hellweg and KalusLang et al, 2005) and another study found an association between the Met allele and lower scores of neuroticism, a risk factor for depression (Reference Sen, Nesse and StoltenbergSen et al, 2003). Thus, it remains unclear whether the association is a direct effect or a confounding factor through some indirect mechanism such as personality traits.
Summary and outlook
In summary, we hope that our findings will shed some light onto the mixed results surrounding the relationship between the BDNF gene and bipolar disorder. According to our findings, the BDNF gene could be interpreted as a genetic risk factor for distinct symptoms found in bipolar disorder and other major neuropsychiatric disorders. We hypothesise that rapid cycling is an important feature in the phenotype associated with the BDNF gene. Furthermore, another study finds this same specific association (Reference Green, Raybould and MacgregorGreen et al, 2006). None the less, our results should be regarded as preliminary and the specificity of the BDNF gene for rapid cycling in bipolar disorder requires replication in further studies. Finally, it is important to bear in mind that effect size of the BDNF gene polymorphism appears to be relatively small, with maximum relative risk factors at about 3 for single marker association tests. This, however, is consistent with other complex disorders, where a variety of genes, all bearing small-to-moderate impact on the genetic susceptibility, are observed. In mood disorders, it is probable that different sets of genes predispose to overlapping phenotypes, some of which belong to the spectrum of bipolar disorder (Reference KelsoeKelsoe, 2003). The relatively consistent association of the BDNF gene with mood disorders should, with more research, lead to improved understanding of their aetiology and may pave the way for novel and more efficient diagnostic and treatment strategies.
Acknowledgements
We thank Matthew Lanktree and Mary Smirniw who assisted in this study. This work was supported by a Canadian Institutes of Health Research operating grant to J.L.K and a postdoctoral fellowship award to D.J.M.









eLetters
No eLetters have been published for this article.