Kashin-Beck disease (KBD) is a disabling osteoarticular disease, which affects people in a crescent-shaped region through Northern China, Mongolia, Siberia and North Korea. KBD often attacks the growth of the joint cartilage, causing deformity of joints, which finally results in dwarfism and joint pain(Reference Tomlinson1). The major pathological changes in KBD patients are chondrocyte necrosis in the hypertrophied layer of the cartilage, with cell cluster formation nearby(Reference Guo2). Although the aetiology and pathogenesis of KBD are still unclear, three hypotheses have been proposed, namely Se deficiency, exposure to fusarium toxins and drinking-water with organic matter (humic acids and fulvic acids)(Reference Tan and Wen3). Some researchers suggest that iodine deficiency(Reference Moreno-Reyes, Suetens and Mathieu4) and viral infection(Reference Bi5) are also risk factors for KBD. Among these potential causes, Se deficiency is ubiquitous in the KBD area, and Se supplementation could significantly decrease the incidence rate and ameliorate disease severity(Reference Li, Li and Hu6, Reference Li, Zhao and Cao7). Se deficiency is considered to be the main environmental factor for KBD. However, a dose–response relationship between low Se and KBD in a prospective study was not observed(Reference Guo, Ding and Zeng8). Furthermore, the incidence of KBD exhibits obvious familial aggregation(Reference Shi, Guo and Ren9). Therefore, it is accepted that the aetiology of KBD is multifactorial and hereditary factors may play a part in the occurrence of KBD.
Several studies(Reference Gao, Zhou and Yu10, Reference Yu, Guo and Gao11) have revealed that gene variance is related to the incidence of KBD. The GPx1 gene may contribute to higher susceptibility to KBD(Reference Xiong, Mo and Zou12). Some research(Reference Regina and Franz13, Reference Antonia, Kostja and Petra14) has shown that Se deficiency is a putative risk factor for osteoarthropathy as selenoproteins have an impact on bone physiology. Therefore, the interaction of susceptibility genes and harmful environment factors may lead to the occurrence and development of KBD.
Glutathione peroxidase 4 (GPx4) is a selenoprotein that directly acts on phospholipid hydroperoxide within cell membranes and lipoproteins. It is also involved in the apoptosis and regulation of gene expression(Reference Imai and Nakagawa15). In selenoproteins, Se is present as an amino acid selenocysteine (Sec) and its incorporation occurs during the synthesis of proteins. This incorporation requires the recording of UGA codon that involves the formation of a complex between an RNA stem-loop structure (Sec insertion sequence) located in the 3′ untranslated region (3′UTR) of selenoprotein mRNA and various trans-acting proteins(Reference Papp, Lu and Holmgren16).
GPx4 expression depends on both dietary Se intake and genetic factors, and variants in the genes that encode GPx4 could influence antioxidant capacity and other functions(Reference Méplan, Crosley and Nicol17), which may change susceptibility to excessive oxidative stress. Several studies(Reference Méplan, Hughes and Pardini18–Reference Maiorino, Bosello and Ursini22) have investigated the association between genetic variation of GPx4 and the increased risk of cancers and other diseases. A common SNP of GPx4 has been found, which is located in the 3′UTR near the Sec insertion sequence element at position 718. GPx4c718t polymorphism (rs713041) could modulate the synthesis of GPx4 by altering the affinity of the Sec insertion machinery for its Sec insertion sequence element and protein binding to the 3′UTR. Some studies(Reference Bermano, Pagmantidis and Holloway19–Reference Maiorino, Bosello and Ursini22) indicate that this locus is associated with lipid metabolism and several cancers. Another SNP (rs4807542) was chosen as a tagging SNP rather than a known functional variant. Both SNP are in the exons that might influence the gene expression. Therefore, it is very important to explore the relationship between GPx4 polymorphism and the risk of KBD. Furthermore, we also detected the mRNA expression of GPx4 in order to explore the pivotal role of GPx4 in the pathogenesis of KBD.
Materials and methods
Study population
According to the national diagnosis of KBD (WS/T207-2010; Ministry of Health, People's Republic of China), 219 patients from KBD areas (Xunyi, Linyou, Qianyang, Long, Changwu and Yongshou counties) were enrolled in the study based on radiographic examination (X-ray of the right hand) and clinical diagnosis (degree I–III). A total of 194 healthy subjects, with no signs or symptoms of joint disease and of matching age and sex, were also selected from the Shaanxi province for control. All subjects in the present study were of the Chinese Han population. The present study was conducted according to the guidelines laid down in the Declaration of Helsinki and all procedures involving human subjects were approved by the Human Ethics Committee of Xi′an Jiaotong University, Shaanxi, People's Republic of China. Written informed consent was obtained from all subjects.
Genotyping analysis
Data from the International Genome Project and resequenced data from the Genome Variation Server (http://gvs.gs.washington.edu/GVS/) was used to identify tagging SNP. Two polymorphic variants of GPx4 (rs713041 and rs4807542) were identified with minor allele frequency >0·05 with a correlation of 0·8 or greater. DNA was isolated from peripheral blood lymphocytes using classical protease K extraction and used for PCR-restriction fragment length polymorphism. The PCR details were as follows: the rs713041 polymorphism was determined in the Eppendorf gradient-type mastercycler (Eppendorf, Hamburg, Germany) with a total volume of 12·5 μl, containing 6·25 μl 2 × Taq PCR MasterMix (RunDe, Xi′an, Shaanxi, People's Republic of China), 0·4 μl each primer (10 μm), 1·5 μl genomic DNA and 3·95 μl double distilled water (ddH2O). PCR was performed at 94°C for 4 min, followed by thirty-five cycles of denaturation at 94°C for 30 s, annealing at 57°C for 30 s and extension at 72°C for 1 min and a final elongation step at 72°C for 10 min. The amplification products were digested at 37°C overnight with a Sty I restriction endonuclease (Takara, Dalian, Liaoning, China). PCR protocols for rs4807542 were the same as rs713041 except the annealing temperature and restriction endonuclease. All the primers used in the study were synthesised by Beijing AuGCT Biotechnology Company, Limited (Beijing, China). The primers, annealing temperature and restriction endonucleases used in study are listed in Table 1.
Table 1 Conditions used for genotyping assays and real-time PCR of glutathione peroxidase 4 (GPx4)
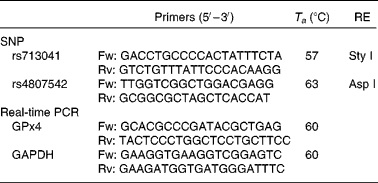
T a, annealing temperature; RE, restriction enzyme; Fw, forward; Rv, reverse; GAPDH, glyceraldehyde 3-phosphate dehydrogenase.
Enzyme-digested products were subjected to electrophoresis in a 2·5 % agarose gel with 0·5 μl ethidium bromide and visualised by a Light Transilluminator (UVP, Upland, CA, USA). In addition, 20 % of the DNA samples were randomly selected for re-genotyping.
Quantitative real-time PCR
A total of thrity-six blood samples, eighteen from each of the KBD patients and healthy subjects, were randomly collected for mRNA quantification. Total RNA were isolated from 3 ml whole blood using the TRIzol reagent (Invitrogen, Carlsbad, CA, USA) according to the manufacturer's instructions, and reverse-transcribed in a thermocycler using the RevertAidTM First Strand complementary DNA Synthesis Kit (MBI, Fermentas, Vilnius, Lithuania) according to the manufacturer's instructions.
Quantification of mRNA of GPx4 was performed on iQ™5 Real-Time PCR Detection Systems (Bio-Rad, Philadelphia, PA, USA) using BioEasy SYBR Green Real Time PCR Kit (Bioer, Hangzhou, Zhejiang, China) according to manufacturer's instructions. The reactions were performed in a 25 μl mixture containing 2 μl complementary DNA, 0·5 μl each primer (10 μm), 12·5 μl 2 × SYBR Mix (with 4·0 mm-Mg2+), 0·15 μl Taq DNA polymerase and 9·35 μl ddH2O using the following program: initial denaturation at 94°C for 2 min, followed by forty cycles of denaturation at 94°C for 10 s, annealing at 60°C for 15 s and extension at 72°C for 30 s. All reactions were performed in duplicate. The results were normalised with the levels of glyceraldehyde 3-phosphate dehydrogenase expression and analysed by iQ™5 software (version 2.0; Bio-Rad) and SPSS 13.0 (SPSS, Chicago, IL, USA).
Statistical analysis
Distribution normality was assessed using the Kolmogorov–Smirnov test. Differences without skewness between the KBD and control groups were measured using an independent sample Student's t test. Deviation of the genotypic frequencies from the Hardy–Weinberg equilibrium (HWE) was assessed in the controls and cases by χ2 test of each locus with 2 df. Categorical variables were presented using frequency counts and compared by a χ2 test. The G*Power program (G*power 3.1.2; http://www.psycho.uni-duesseldorf.de/abteilungen/aap/gpower3/) was used to estimate the statistical power under a two-tailed test. The present sample size showed 79 % power to detect significance (α < 0·05) in the association with allele, genotype under study conditions and an effect size index of 0·2 (corresponding to ‘weak’ gene effect) was used. The relative risks of KBD were measured as OR with 95 % CI. SHEsis software (http://analysis.bio-x.cn/myAnalysis.php) was used to analyse the linkage disequilibrium (LD) measurements (D′ and r 2) and construct haplotypes. Other statistical analyses were calculated with SPSS 13.0 software. P < 0·05 was considered statistically significant (two-sided).
Results
Baseline characteristics
A total of 219 KBD patients and 194 controls with frequency matched by age and sex were included in the study. In addition, KBD patients were divided into I, II and III degrees according to clinical diagnosis. The baseline characteristics are shown in Table 2. Sex and age of control and KBD group had no significant difference (P = 0·162 and P = 0·736, respectively).
Table 2 Characteristics of the study population
(Mean values and standard deviations or percentages)
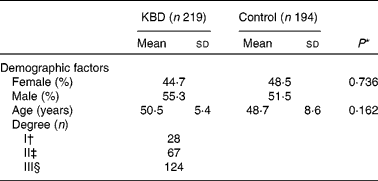
KBD, Kashin-Beck disease.
* Mean values were significantly different (P < 0·05).
† Diagnosis of KBD (WS/T 207-2010): multiple, symmetrical enlargement of fingers. Restricted movement, enlargement, pain and mild amyotrophy in other limb joints.
‡ Diagnosis of KBD (WS/T 207-2010): shortened fingers and aggravated clinical symptoms and signs of first degree.
§ Diagnosis of KBD (WS/T 207-2010): retarded growth or dwarfism and aggravated clinical symptoms and signs of second stage.
Genotype analysis
In the present study, we selected two tagSNP in exons of GPx4, which were highly polymorphic in both the groups. A departure from the genotype distribution was found in the KBD cases of rs713041 (P = 0·0036), while the others were in HWE (P>0·05), a trend test was performed subsequently.
Table 3 shows that genotype and allele frequencies of the two SNP for KBD patients and control subjects in Han Chinese. No significant differences in genotype distribution and allele frequencies were found in the controls and the KBD cases. The same results were achieved after performing the trend test. In addition, we compared the genotype and allele frequencies of the two SNP of the control group in the Chinese Han population with data published from the HapMap project. As presented in Table 3, genotype frequencies of both SNP in the control group were consistent with the data from HapMap, suggesting that the restriction fragment length polymorphism-PCR was a reliable method for examining gene variation.
Table 3 Genotype and allele frequencies of polymorphisms between Kashin-Beck disease (KBD) and control groups†
(Numbers, percentages, odds ratios and 95 % confidence intervals)

P t, P value for trend test; CHB, Han Chinese in Beijing, China (HapMap representative chinese population).
* Mean values were significantly different (P < 0·05).
† Categorical variables were presented using frequency counts and compared by χ2 test.
‡ Data from HapMap.
The LD analysis was further performed (noted as D′ and r 2) between the two SNP for both groups. D′ was 0·662 while r 2 was 0·053. Therefore, medium LD was observed in the two SNP LD estimation (D′>0·6).
Both SNP were constructed in four haplotypes (shown in Table 4). Individual haplotype frequencies were compared between KBD cases and controls, and a significant difference was observed in the haplotype AT (P = 0·0066), which was significantly lower in the KBD cases compared with the controls (0·006 v. 0·032 %). When the distributions of the haplotypes were compared, we found significant differences between KBD cases and controls in the haplotype combination of rs713041 and rs4807542 (χ2 = 9·98, global P = 0·019).
Table 4 Loci chosen for hap-analysis: rs4807542, rs713041†
(Numbers, percentages, odds ratios and 95 % confidence intervals)
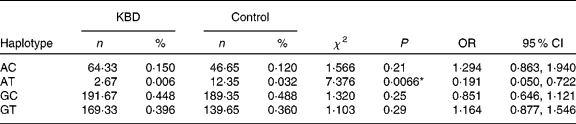
KBD, Kashin-Beck disease.
* Mean values were significantly different (P < 0·01).
† As the haplotype frequency is estimated by program, the accurate results are not integer numbers in most of the cases.
Glutathione peroxidase 4 mRNA expression
The GPx4 mRNA level was assessed by real-time PCR and glyceraldehyde 3-phosphate dehydrogenase complementary DNA was used for normalising GPx4 expression. Whole blood GPx4 mRNA expression in the KBD group was significantly lower compared with the controls (0·058-fold, P < 0·001, n 18).
Discussion
KBD is an endemic joint disease, which is mainly distributed in areas with low Se intake. So far, the aetiology of KBD is still obscure. Except three major aetiology hypotheses, some researchers also proposed other risk factors, such as iodine deficiency(Reference Moreno-Reyes, Suetens and Mathieu4) and viral infection(Reference Bi5). Moreno-Reyes et al. (Reference Moreno-Reyes, Mathieu and Boelaert23) reported that iodine and Se deficiency coexist in KBD areas and iodine supplementation had some effect on KBD patients in Tibet(Reference Moreno-Reyes, Mathieu and Boelaert23). Furthermore, animal experiments showed that iodine deficiency could aggravate Se deficiency(Reference Su, Zhang and Tian24). However, epidemiological studies indicate that KBD is not always found in iodine deficiency areas but in Se deficiency areas. Recently, parvovirus B19 have also been found in the peripheral blood of KBD patients(Reference Xiong, Wang and Dai25), but whether it plays a role in KBD aetiology needs to be further confirmed. Therefore, Se deficiency may be the main environmental factor for the occurrence of KBD. KBD is focally distributed and shows a clustering in families(Reference Shi, Guo and Ren9), Furthermore, Se supplementation could not entirely prevent KBD, which suggests that some additional mechanism might be playing a role in KBD development.
GPx4 is an antioxidative selenoprotein extinguishing lipid peroxide in mammals and also involved in other cell functions such as apoptosis and gene expression regulation(Reference Imai and Nakagawa15). Several studies on the polymorphism of GPx4 showed that gene variance might influence GPx4 expression and activity. Considering that gene variation might influence the susceptibility of KBD, we performed this case–control study to explore the relationship between polymorphism of GPx4 and KBD.
The results showed no differences in either genotype or allele frequency of SNP rs713041 and rs4807542 between healthy and KBD subjects. Meanwhile, single-locus association analysis for the two SNP was performed and an obvious LD was observed. Furthermore, we constructed four haplotypes, and the analysis of haplotype distribution revealed that the combination of two SNP showed a potential association with KBD (global P = 0·019). When each haplotype was compared in the KBD cases and controls, a significant difference was found in haplotype A-T (P = 0·0066). The results indicated that A-T haplotype may be a protective factor for KBD. Thus, a functional locus that affects the risk of KBD might exist in GPx4, which further implied that GPx4 might be a susceptibility gene for KBD. Specifically, rs713041 is in the 3′UTR, which plays an important role in eukaryotes for the following reasons: (1) 3′UTR is important for regulating selenoprotein gene expression in determining the relative extent to which the different mRNA are translated. (2) Differences in the 3′UTR sequences are also believed to be responsible for the differences in the relative translation rates and mRNA stability(Reference Hesketh and Villette26). In addition, rs713041 has been shown to be associated with lymphocyte 5-lipoxygenase metabolites(Reference Villette, Kyle and Brown21). One possibility is that the alteration in GPx4 leads to change in 5- and 15-lipoxygenase activity, cyclo-oxygenase-2 activity and activation of the NF-κB pathway(Reference Imai and Nakagawa15), reflecting the role of GPx4 as a regulator of the inflammatory responses.
The distribution of the SNP in the control group was compared with the HapMap project of the Chinese Han population. The results showed a similar distribution in either genotype or allele frequency of both SNP. Therefore, the control samples in the present study were representative.
No differences were found in either genotype or allele frequency of SNP rs713041 and rs4807542 between the KBD patients and healthy subjects. This result should be interpreted with caution because a departure from HWE for rs713041 was observed in the KBD cases (P = 0·0036). Zou et al. (Reference Zou and Donner27) recently proposed that a trend test should be performed for the data deviating from HWE. Therefore, we recalculated the data using the trend test and no significant differences in KBD samples (P = 0·839) were found, suggesting that the departure of HWE imposed little impact on our result.
GPx4 was an extremely stable enzyme, which was preferentially spared in Se-depleted animal models indicating that it was a marker for reflecting the status of Se(Reference Sunde and Hadley28, Reference Brown, Pickard and Nicol29). Furthermore, GPx4 mRNA levels were high in the blood(Reference Evenson, Wheeler and Blake30). We investigated the mRNA expression of GPx4 in the present study. The result showed that the mRNA level in the whole blood was significantly lower in KBD group compared with the controls (P < 0·001; Fig. 1), suggesting that KBD patients were Se deficient. GPx4 was known as the major antioxidant against lipid peroxide, under the condition of GPx4 decreased, lipid peroxide metabolism balance would be disturbed, which could explain the excessive lipid peroxide and NEFA in the serum of KBD patients(Reference Guo2).

Fig. 1 Glutathione peroxidase 4 (GPx4) mRNA level in whole blood in Kashin-Beck disease (KBD, ![]() ) and control (
) and control (![]() ) groups. GPx4 mRNA level of whole blood was 0·058-fold lower in the KBD than in control group. Values are means, with their standard deviations represented by vertical bars (n 6, P < 0·001). * Mean values were significantly different (P < 0·01).
) groups. GPx4 mRNA level of whole blood was 0·058-fold lower in the KBD than in control group. Values are means, with their standard deviations represented by vertical bars (n 6, P < 0·001). * Mean values were significantly different (P < 0·01).
In summary, the haplotypes composed of rs713041 and rs4807542 suggested that A-T may be a protective haplotype against Se-deficient environment. GPx4 polymorphisms and the low expression of mRNA may be related to the development of KBD in the Chinese population. This was the first study on the association between GPx4 polymorphism and the risk of KBD. However, the sample size in the present study is relatively small; further clinical and experimental investigations are needed to confirm the present conclusion.
Acknowledgements
The authors have no conflict of interest and certify this to be a true and original study. The study was supported by grants from National Science Foundation of China (no. 30671820) and the Natural Science Foundation of Shaanxi Province, China (grant no. SJ082T12-5). X. H. D. carried out the studies and drafted the manuscript. R. X. S., X. Z. Z., W. Y. S. participated in design and statistical analysis, and sample collection. G. L. B. contributed to sample collection. X. X. D. and X. Y. M. participated in study design and helped to draft the manuscript. Y. M. X. conceived the study, participated in design and assisted with the manuscript. We are deeply thankful to all the volunteers who participated in the present study, as well as to Jian Hong Zhu (The Second Affiliated Hospital, Peoples Republic of China) and Yue Xiang Yu (Shaanxi Provincial Institute for Endemic Disease Control, Peoples Republic of China) for their assistance in sample collection.







