Published online by Cambridge University Press: 03 September 2021
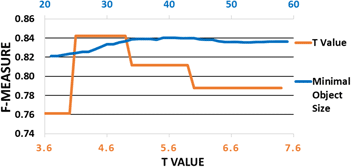
Cell confluence is an important metric in cell culture, as proper timing is essential to maintain cell phenotype and culture quality. To estimate cell confluence, transparent cells are observed under a phase-contrast or differential interference contrast microscope by a biologist, whose estimations are error-prone and subjective. To overcome the necessity of using the phase-contrast microscope and reducing intra- and inter-observer errors, we have proposed an algorithm that automatically measures cell confluence by using a commonly used brightfield microscope. The proposed method consists of a transport-of-intensity equation-based brightfield microscopic image processing, an image reconstruction method, and an adaptive image segmentation method based on edge detection, entropy filtering, and range filtering. Experimental results have shown that our method has outperformed several popular algorithms, with an F-score of 0.84 ± 0.07, in images with various cell confluence values. The proposed algorithm is robust and accurate enough to perform confluence measurement with various lighting conditions under a low-cost brightfield microscope, making it simple and cost-effective to use for a fully automated cell culture process.