Methicillin-resistant Staphylococcus aureus (MRSA) is a common hospital pathogen worldwide and may be the cause of severe infections. Its multiple antibiotic resistance poses a therapeutic challenge and patients with MRSA tend to have a significantly longer in-hospital stay, worse prognosis and higher mortality rate. These patients also represent higher costs to the health system, and may be subject to social stigma and greater psychological stress [Reference Cosgrove1].
Several genotype and phenotype analysis studies have confirmed that some MRSA strains become endemic within the hospital environment, and specific epidemic clones have emerged with enhanced ability to spread within and among hospitals and to cross national boundaries [Reference Oliveira2]. A relatively small number of pandemic MRSA clones cause the majority of MRSA infections. Multilocus sequence typing and SCCmec-type analysis of isolates from Southern Europe, the USA, and South America, showed that nearly 70% of these isolates belonged to five major pandemic clones, i.e. Iberian (ST 247-MRSA-IA), Brazilian (ST239-MRSA-IIIA), Hungarian (ST239-MRSA-III), New York/Japan (ST5-MRSA-II), and Pediatric (ST5-MRSA-IV) clones [Reference Enright3]. In addition, the EMRSA-15 (ST22-IV), and EMRSA-16 (ST36-II) clones, are dominant in England and Scotland [Reference Johnson4].
According to different studies, MRSA clones disseminated in South America belonged mainly to the Brazilian Epidemic Clone (BEC) (ST239-MRSA-IIIA) which was identified in Argentina, Brazil, Chile, and Uruguay between 1992 and 1998 [Reference Rodriguez-Noriega5]; clone (ST5-MRSA-IV) was identified in Colombia between 1996 and 1998 [Reference Oliveira6], and the Cordobes/Chilean clone in Chile and Argentina between 1998 and 2002 [Reference Sola7, Reference Sola8]. In Brazil, BEC is widespread in hospitals [Reference Teixeira9] and in Rio de Janeiro during 1999–2000 it was shown to coexist with strains resembling the Hungarian clone and the Pediatric clone [Reference Vivoni10]. The BEC has also been detected as the cause of invasive infection in Argentina and Uruguay [Reference Da Silva Coimbra11] but the molecular epidemiology of MRSA in Latin America is still largely unknown.
The objective of this study was to evaluate the molecular epidemiology of MRSA involved in nosocomial infections in Porto Alegre city and its metropolitan area in southern Brazil with a population of about four million people. From April to December 2008 patients with MRSA infections at the city and peripheral hospital (each with about 100 beds) were studied. Clinically and epidemiologically relevant information from each patient was collected from the medical records and the first isolate only from each patient was subjected to further study. Susceptibility to antimicrobial agents was determined by minimal inhibitory concentrations (MICs) on the MicroScan WalkAway system (Siemens Healthcare, USA) according to the protocols of the Clinical and Laboratory Standards Institute [12].
Methicillin resistance was confirmed by amplification of an internal fragment of the mecA gene by PCR [Reference Vannuffel13] and SCCmec typing was performed as previously described [Reference Zhang14]. SCCmec-type control strains were NCTC10442 (I), N315 (II), 85/2082 (III), CA05 (IVa), 8/6-3P (IVb), MR108 (IVc), JCSC4469 (IVd) and WIS (V).
Analysis of chromosomal DNA of MRSA isolates was performed by pulsed-field gel electrophoresis (PFGE), according to the Centers for Disease Control and Prevention (CDC, USA) protocol for S. aureus [Reference McDougal15]. Gels were normalized with reference strain S. aureus NCTC 8325 and compared to representative strains of local and global MRSA clones: A1721/HU25 (BEC), WB72 (USA300), MW2 (USA400), WB49 (Oceania South Pacific clone), HAR24 (EMRSA-15), BK2464 (New York/Japan clone), and HDE288 (Pediatric clone/USA800). DNA profiles were interpreted by visual inspection and by the unweighted pair-group method using arithmetic averages (UPGMA) based on Dice coefficients with Bionumerics software, version 5.0 (Applied-Maths, Belgium). Strain relatedness was displayed as a dendrogram and a similarity coefficient of 80% was used to distinguish between lineages [Reference McDougal15].
Thirty S. aureus isolates from inpatients were resistant to methicillin. These patients showed a broad array of underlying clinical conditions, which included intravascular infection (27%), surgical interventions (33%), and respiratory disorders (24%); 15% had no underlying conditions (15%). All patients had a history of hospitalization for long periods and fulfilled criteria for hospital-associated infection. Isolates were recovered from surgical and skin wounds (36%), respiratory tract (24%), blood (15%), intravascular devices (12%), biopsy (6%), and ascitic fluid (6%).
All MRSA isolates were positive for the mecA gene by PCR. Most (⩾90%) were multi-resistant and exhibited resistance to ciprofloxacin, clindamycin, erythromycin, and gentamicin; 7/30 (23%) were resistant to trimethoprim-sulfamethoxazole and two isolates to rifampicin. All were susceptible to linezolid and vancomycin. Eighteen isolates had PFGE profiles clonally related to BEC and carried SCCmec type III, and 11 isolates showed PFGE profiles highly similar to the Cordobes/Chilean clone and harboured SCCmec type I; the remaining isolate was distinct from these clones and was of SCCmec type IV (Fig. 1). Isolates of SCCmec type I were typically multidrug resistant (except for trimethoprim-sulfamethoxazole and rifampicin), whereas those with SCCmec type III had a variable pattern of resistance (Table 1).
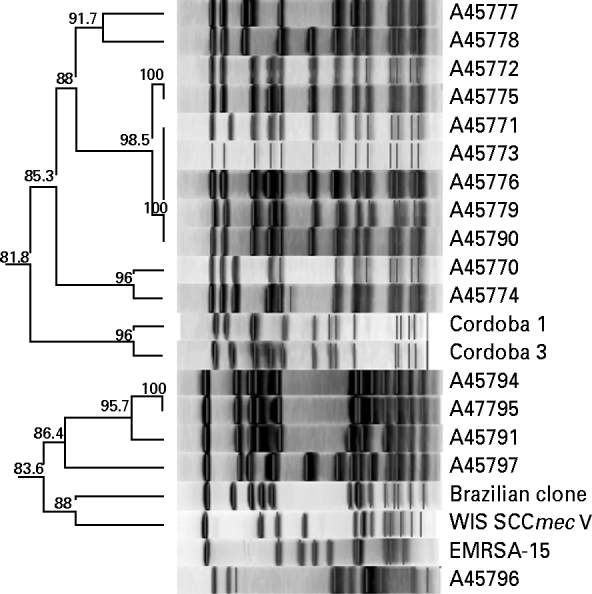
Fig. 1. Cluster analysis of percentage similarities of MRSA clinical isolates compared to international reference standard clones. A similarity coefficient of 80% was selected to distinguish between clusters.
Table 1. Antimicrobial resistance of MRSA isolates according to SCCmec type
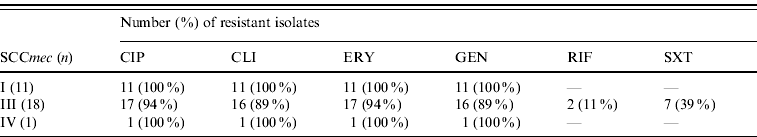
CIP, Ciprofloxacin; CLI, clindamycin; ERY, erythromycin; GEN, gentamicin; RIF, rifampicin; SXT, trimethoprim-sulfamethoxaole.
Relatively few clones of MRSA are distributed worldwide. The Brazilian clone and variants of it continue to circulate throughout hospitals in Brazil as well as neighbouring countries. However, several other MRSA clones have successfully spread in particular regions of the country, especially those related to the New York/Japan and the Pediatric clones [Reference Rodriguez-Noriega5]. The Cordobes/Chilean clone was first identified in isolates from Chile in 1997, and in Argentina in 1999; it quickly became predominant over the Brazilian clone in Argentina in 2001 (53% vs. 23% in hospital-acquired infections). [Reference Sola8] Such a replacement of a specific epidemic MRSA clone by another has been observed in several other countries, such as the Czech Republic [Reference Melter16], Greece [Reference Aires de Sousa17], and Mexico [Reference Velazquez-Meza18].
The Cordobes/Chilean clone has not been described so far in Brazil and previous comprehensive studies did not detect MRSA SCCmec type I among Brazilian isolates [Reference Oliveira6, Reference Soares19, Reference Sousa-Junior20]. Recently, isolates of SCCmec type I were detected in another hospital of Porto Alegre but a clonal analysis was not presented [Reference Santos21]. The question of when and how transmission of the clone occurred between the two countries cannot be answered by this study but as Brazil and Argentina share a long border, patients may receive hospital care in both countries. Moreover, tourism and trade are intense between these two countries.
The susceptibility of the Cordobes/Chilean clone to trimethoprim-sulfamethoxazole could be a useful phenotypic marker in the clinical microbiology laboratory to differentiate this from the Brazilian clone in a suspected outbreak [Reference Sola7] and consistent with that all isolates of the former clonal lineage were susceptible to this agent. Clinical and epidemiological data retrieved from patient medical records did not show any association between clonal group and type of infection, ward of isolation, or group of patients. All patients remained hospitalized for long periods and fulfilled criteria for hospital-associated infection. Most had a history of previous surgery and respiratory disorders and the remainder other varied underlying conditions; however, larger more focused studies are necessary to compare clinical predisposition and outcome associated with the two clones.
We have shown the presence and potential dissemination of the Cordobes/Chilean clone in Porto Alegre, Brazil which possibly could displace the currently dominant BEC. Future follow-up surveillance studies of the molecular epidemiology of MRSA in Brazilian hospitals are therefore of crucial importance to inform infection control measures and reduce the impact of these infections in hospital patients.
ACKNOWLEDGEMENTS
We thank K. Hiramatsu and T. Ito for the kind gift of SCCmec control strains and R. S. Daum, H. de Lencastre and A. Figueiredo for other reference control strains. We also thank LEMC (Laboratório Especial de Microbiologia Clínica – Unifesp) in particular Professor Antonio Carlos de Campos Pignatari for support in performing PFGE.




